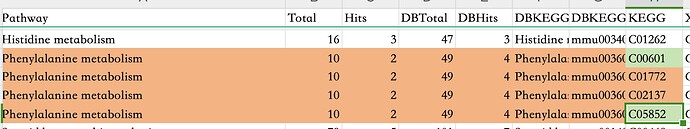
I used MetaboAnalyst 6.0 for pathway analysis and cross-checked the results using the KEGG API. I found that while MetaboAnalyst 6.0 identified some compounds (highlighted in green in the attached image) for a specific pathway, the KEGG API confirmed the presence of two additional compounds relevant to the pathway that were not detected by MetaboAnalyst 6.0. Could this discrepancy be attributed to differences in database versions? Furthermore, would this discrepancy impact the calculation of impact and p-values?
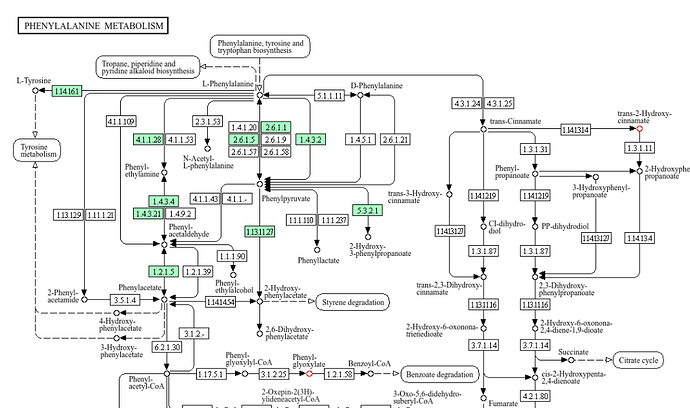
Hello! The pathways curated by MetaboAnalyst are organism-specific. As an example, in the case of phenylalanine metabolism, the KEGG pathway contains 49 compound entries, however, human-specific pathway will only contain the portion where the enzymes are highlighted in green, meaning they are present in the chosen organism and typically have experimental validation. The two compounds C01772 and C02137 (red circles in pathway) are not present in the pathway analysis result for this reason.
Hope it helps!