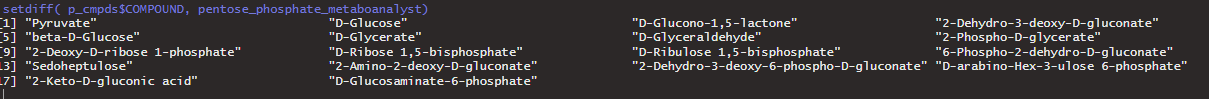Hi all,
I’m trying to perform Pathway Analysis in Metaboanalyst on sorghum samples. We are selecting the maize pathway library but when we get to the results page, some IDs in our input (entered as KEGG IDs) don’t show up in pathways at all, despite knowing they are present in the KEGG pathway for maize.
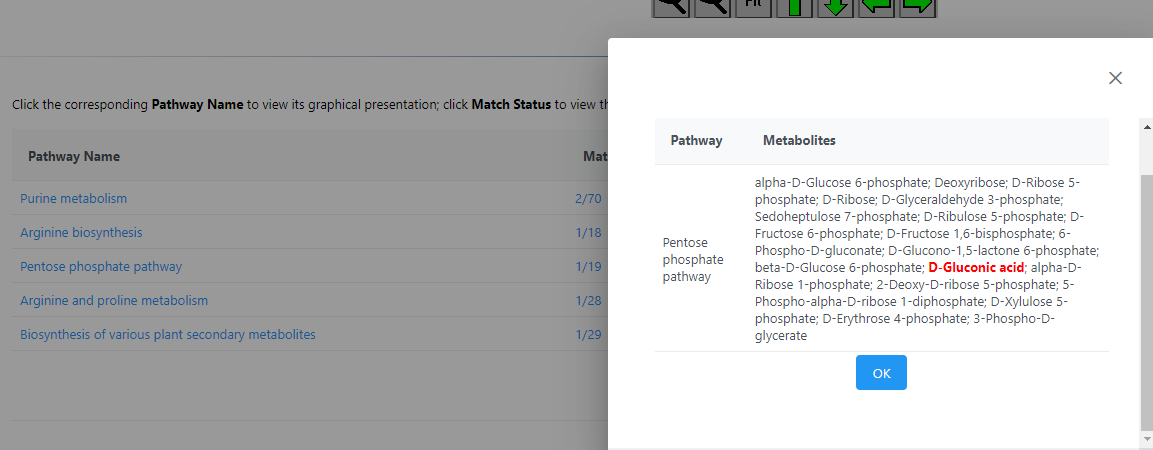
For example, here is what is included in the MetaboAnalyst version of the Pentose Phosphate Pathway (19 compounds):
Here is everything in the KEGG Pentose Phosphate Pathway of Maize that is NOT in the Metaboanalyst (37 compounds) :
So the question is, if these pathways are sourced from KEGG, why are they not all there? This in issue with all the pathways I have checked, not just PPP. All the IDs are recognized by MetaboAnalyst when entered. All have KEGG IDs, HMDB, and PubChem IDs.
On the parameter setting page before the results, it says KEGG pathways were last obtained Dec 2023. To me this isn’t an updating issue.
Any ideas??