Hi there,
Regarding the 44 metabolite sets related to disease signatures found in fecal samples, what’s the relationship between the metabolites in each set and the disease. For instance, among the 93 metabolites in the set for IBS, which are positively correlated with IBS, and which are negatively correlated with IBS. Same question for the other 43 metabolite sets and diereses. Thank you.
Can you clarify which module you are using, and which steps you have taken?
It sounds like you might be referring to the network module, which contains a database relating metabolites, genes, and diseases. There are more details on the databases here.
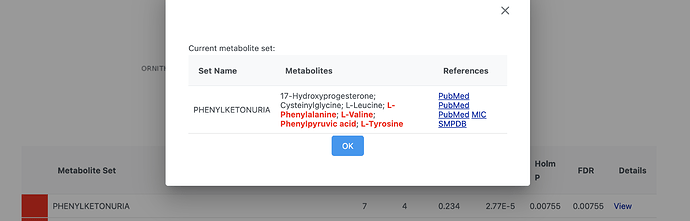
Thank you. It is in the Enrichment analysis, which “44 metabolite sets reported in human feces.” can be chosen.
A table will be obtained in the result ,such as
Metabolite Set Total Hits Expect P value Holm P FDR
Irritable Bowel Syndrome 93 11 1.1 2.01E-10 8.63E-9 8.63E-9
Unclassified Ibd 84 9 0.994 5.87E-8 2.47E-6 8.54E-7
Myalgic Encephalomyelitis/chronic Fatigue Syndrome 22 6 0.26 5.96E-8 2.47E-6 8.54E-7
My question is, for instance, for IBS, among the 93, which are positively correlated with IBS, and which are negatively correlated with IBS. Same question for the other 43 metabolite sets and diereses.
The metabolite sets are signatures curated base on literature - significant metabolites based on their p values (or other criteria reported by authors). We don’t have original data to re-compute them in a consistent way. You may get those details by clicking the “View” link of the metabolite sets to see if the authors have provided such information (see below).