Hello,
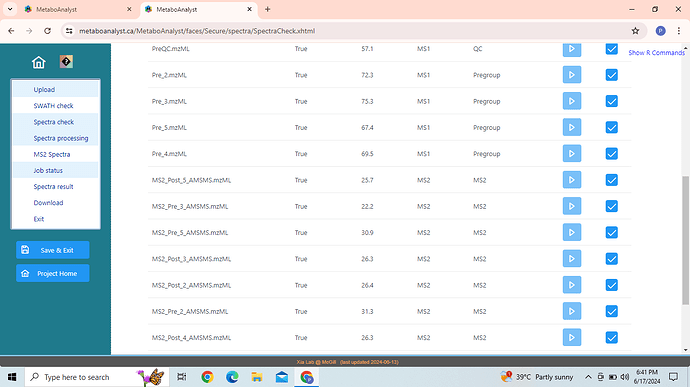
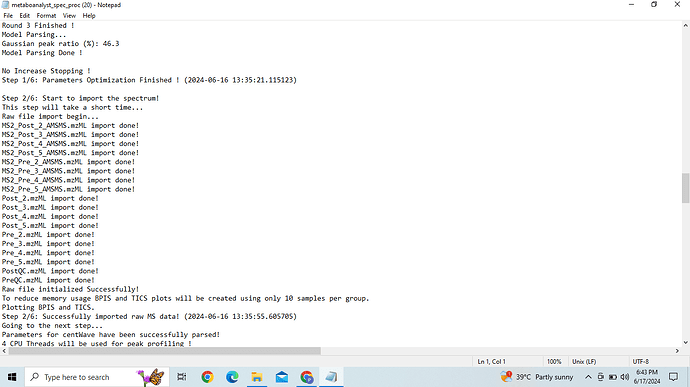
I am using web version of MetaboAnalyst 6.0 since few months now. I am trying to do MS1 + DDA analysis. I have uploaded the individual zip file for MS1 and MS2 spectra along with the Metadata as suggested. Metadata is formatted correctly and Spectra check is done successfully. MS1 and MS2 files are well defined in spectra check. Going further with the spectra processing it also takes MS2 files for MS1 spectra processing which should not be the case as per the tutorial and during the last step of MS1 spectra processing it shows error stating that " No metadata provided, all files in one sub-folder will be considered a group!" I am unable to crack the reason why is this happening and further in MS2 spectra processing it gives error “data is too long” , execution halted.
Even after following all the steps given in the tutorial , things are not working in a way it should be.
Request you to guide me on this @xia.lab @Pang
Regards,
Pooja Shelat
Ph.D scholar , Gujarat University