Hi
I am a new MetaboAnalyst user, and I really like it.
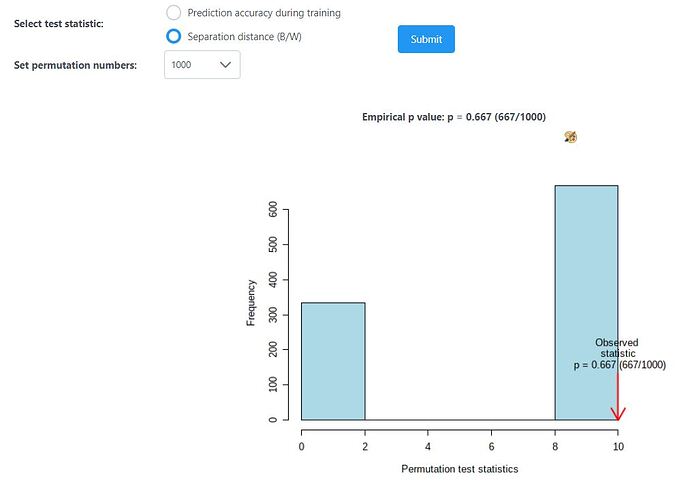
I have a data set containing 2 groups (n-3 per group). Both PCA and PLSDA analysis shows clear clustering of the samples into the groups. However, permutation analysis (B/W with 1000 sets) yields a empirical p value of 0.6. Does this indicate that the separation observed in PLSDA is not real or is the p value high only because the sample size is low (n=3 per group).
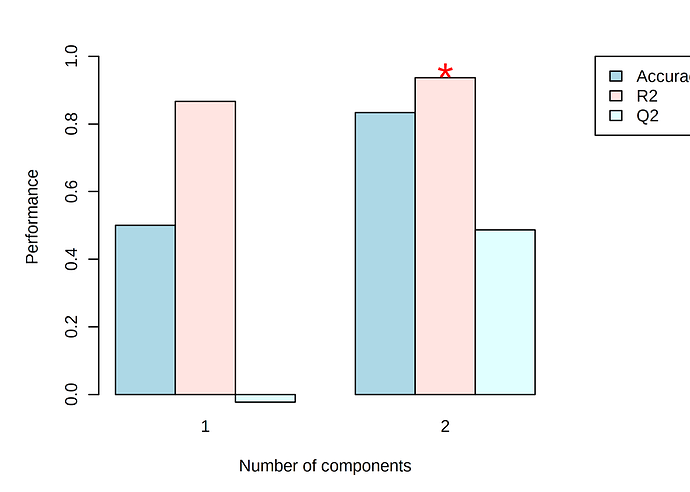
On the other hand, cross validation analysis yields an accuracy of 0.8, Q2 of 0.5 and R2 of 0.9. Is it worth going ahead with the cross-validation results?
pls_score2d_0_dpi72_final.pdf (15.6 KB)
For most datasets analyzed in MetaboAnalyst, the PLS-DA results should be taken as exploratory, to identify potential patterns in the dataset. For a deeper understanding of the permutation p-value in the PLS-DA module, see this post.
Omics datasets often have very low sample size (n = 3 is especially low), where the statistical power is quite low for moderate effect sizes, even if though many of these moderate effect sizes could still be biologically relevant. You can combat this by looking from the data from multiple perspectives (ie. differential abundance analysis, pathway analysis, PCA, PLS-DA, etc) and looking for consistent trends.
The bottom line: prediction / classification (i.e. PLS-DA) generally requires more samples than statistical comparison (i.e. t-tests). Cross validation or permutations are not meaningful with n=3