Hi,
This is a follow-up to my last question: How to do a Joint Pathway Analysis for Candida albicans multi-omics data?
Could you please let me know when I can expect the release of this updated feature?
Thanks.
Hi,
This is a follow-up to my last question: How to do a Joint Pathway Analysis for Candida albicans multi-omics data?
Could you please let me know when I can expect the release of this updated feature?
Thanks.
It was released last Wednesday. Please take a look.
Likely you were re-directed to one of our backup nodes (running the previous version) at the peak time.
Please check your URL and make sure it is: https://www.metaboanalyst.ca/MetaboAnalyst/upload/PathUploadView.xhtml
Thanks.
I tried with the new module. I used gene symbols and compound names as the input, and selected Candida albicans as the organism. I got the following error:
Please follow our post guideline.
Module used: Joint Pathway Analysis
Data: 2 .csv files attached. Proteomics data: UniProt ID, Gene symbol (not available for all, as some Candida proteins are predicted/uncharacterised), Fold change; Metabolomics data: KEGG ID, Fold change
Steps followed:
a. Opened the MetaboAnalyst 6.0 website and selected the Joint pathway analysis tool. Targeted (compound list) was selected from the dropdown menu.
b. In the two dialogue boxes given, first i pasted the list of uniprot IDs with the corresponding fold changes in the proteomics box, and the list of KEGG compound IDs with corresponding fold changes in the metabolomics box.
c. I selected C. albicans as the organism, and clicked on Proceed, getting the same error as before:
d. Since using uniprot IDs didn’t work, i tried the same process with gene symbols only to get the same error again.
Please look into this and help me out.
Thank you!
C.albicans_JointPathwayAnalysis_Metabolomics.csv (952 Bytes)
C.albicans_JointPathwayAnalysis_Proteomics.csv (2.3 KB)
Thank you for the detailed note. I can trace the issue to a typo in the name for this organism - making it never matches the organism code used in the pathway library. It is fixed but will take 24~48 hours for server update
Thank you. The analysis worked with the default parameters. However, i got this error when i tried ‘Combine p values (pathway-level)’ as the Integration method. Can you please resolve this too?
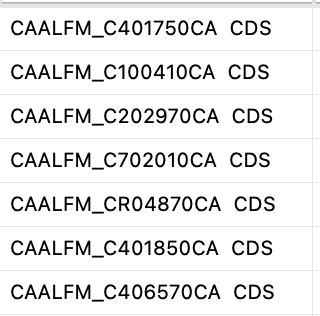
For some organisms, their KEGG gene IDs contain extra text at the end (see below). We have added an extra step to remove this text if found. I have included your data as an example to help us test these special cases in future updates. I hope this is OK with you. You can evaluate after next server update
This topic was automatically closed after 2 days. New replies are no longer allowed.