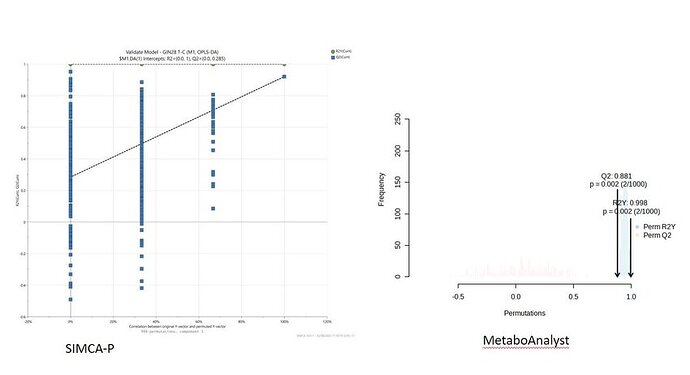
I am doing the permutation test for the OPLS-DA models. I compared two platforms: MetaboAnalyst 5.0 and SIMCA-P using the same dataset. The results from SIMCA-P indicated that there was risk of model over-fitting. However, the results from MetaboAnalyst showed that the empirical p value(Q2 and R2Y) were all less than 0.05. Actually, I am not sure whether the criteria 0.05 I set is correct. How to evaluate my model based on the two different results?
You are comparing an open-source model (k-opls) with a commercial proprietary model (SIMPCA-P). It is unclear whether they are identical, and also the permutation procedure could be different. As for k-opls in MetaboAnalyst, the result is more reliable for two-group evaluation than for multi-group scenario.