I ran a quantitative enrichment analysis (QEA) using MetaboAnalyst, with amino acids and a quantitative phenotype. To my understanding, this analysis uses the globaltest algorithm with a library such as SMPDB.
I also ran a pathway analysis with the same data and saw this analysis also uses the globaltest algorithm with a library such as SMPDB.
I’m having difficulty understanding the difference between these two analyses.
Thank you very much
Hadar Klein
Hi Hadar,
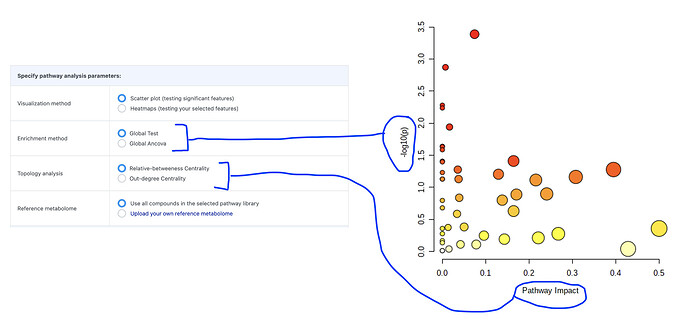
Thanks for the good question. The global test algorithm is the same in the “Enrichment Analysis” and “Pathway Analysis” modules. The difference between them is that for pathway analysis, we do a pathway topology analysis in addition to the global test.
A metabolite set is just a group of metabolites, for example a list of metabolites associated with cancer. A pathway includes both a list of metabolites, and information about how the metabolites interact, for example metabolite A and metabolite B react to form metabolite C in the “Glycolysis” pathway. We can only do enrichment analysis for metabolite sets; we can do both enrichment (analyze metabolites) and topology analysis (analyze metabolites and their connections) for pathways:
Jessica