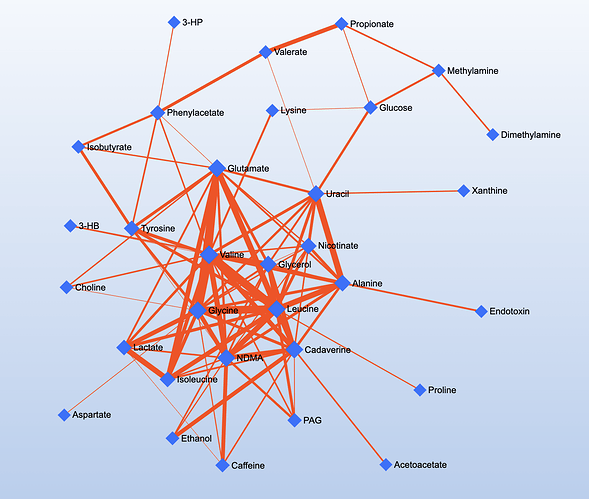
DSPC stands for debiased sparse partial correlation. It is an algorithm designed to build partial correlation networks based on a graphical LASSO model using a data-driven approach without mapping to any known metabolites interaction databases (Jankova, 2015).
DSPC can distinguish between direct and indirect associations and provide insights into the structure of dependencies between metabolites. The main assumption of this DSPC method is that the amount of real metabolites interactions is far smaller than the sample size (i.e., the real partial correlation network among the metabolites is sparse).
Under this assumption, DSPC reconstructs a network and calculates partial correlation coefficients and p-values for each pair of metabolic features. Thus, DSPC makes it possible to uncover connectivity patterns among a large number of metabolic features by using fewer samples (Basu et al., 2017). The inferred results can be visualized as weighted networks, where nodes represent the metabolic features, and the edges depict the correlations among them.
An example DSPC network generated by MetaboAnalyst. Users can manually interact with the networks and manually re-arrange nodes and edges