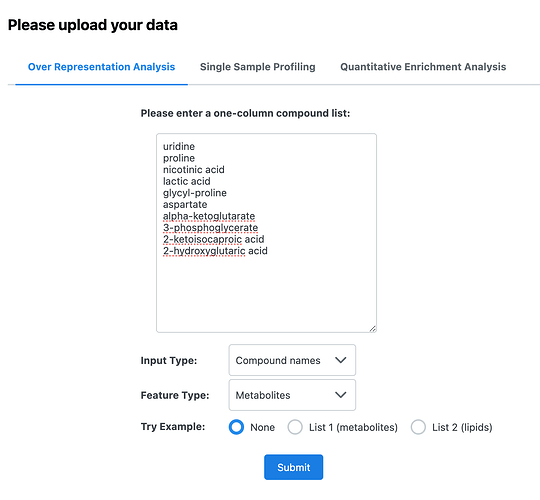
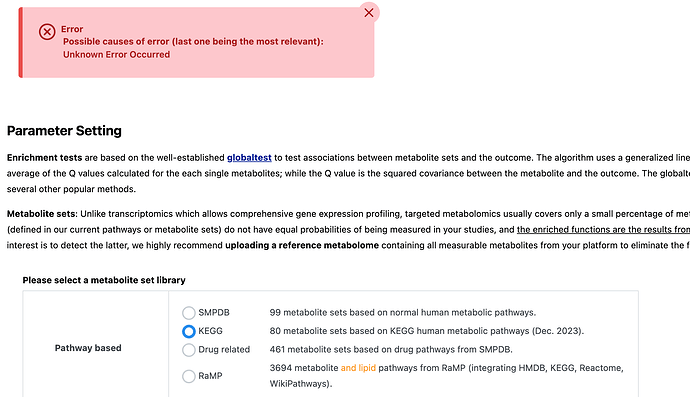
I am trying to conduct a KEGG Pathway Analysis for nine metabolite lists. I was able to conduct the chemical class enrichment for all nine lists. However, I was unable to conduct the analysis for two of these lists due to an “unknown error.”
List 1 (error occurs): https://github.com/yaaminiv/green-crab-metabolomics/blob/main/output/05-metabolomics-analysis/metaboanalyst/module6_metabolites.csv
List 2 (error occurs): https://github.com/yaaminiv/green-crab-metabolomics/blob/main/output/05-metabolomics-analysis/metaboanalyst/module8_metabolites.csv
List 3 (error does not occur): https://github.com/yaaminiv/green-crab-metabolomics/blob/main/output/05-metabolomics-analysis/metaboanalyst/module7_metabolites.csv
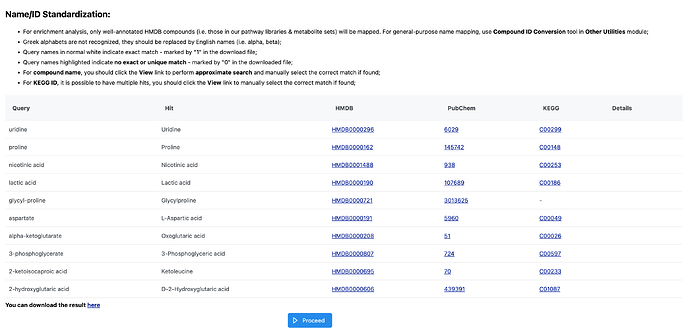
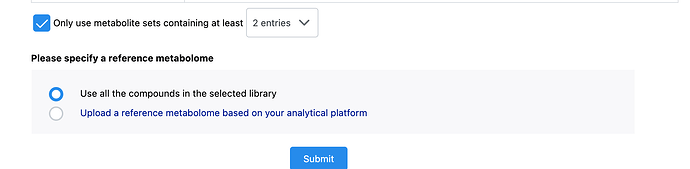
I copy/paste the list of metabolite names for the Over Representation Analysis and proceed after Name/ID Standardization. I select “KEGG” for a Pathway-based enrichment using metabolite sets with at least two entries and compounds in the selected library as the reference. When I click “Submit” for the “Parameter Setting” step, that is when I get the error.
Is there a different way I should be uploading these lists? I’ve tried using KEGG IDs instead of metabolite names and I still get the same error. I was able to perform a Main Compound Class enrichment test with these same lists on MetaboAnalyst and I did not get any errors.