When completing a 2-way ANOVA on a multiple factors/co-variates dataset, following normalization of the dataset it throws an unknown error. When no normalization is done it allows to 2-way ANOVA??
@jess.ewald I am having the same issues. When I updloud my data for 2way ANOVA, I’m getting an “unknown error” aswell. I can see a nice PCA plot, but I cannot get the 2 way ANOVA to work without this unknown error, even after relabeling all compounds, groups and samples based on the example data. See below for all the details:
- Which tool and which module
MetaboAnalyst 5.0 with Statistical Analysis (metadata table) - Provide a copy of your data, or indicate which example data you used
Please see attached:
20221207_Heart_MetaboAnalyst_Input_2Way _Ti_Tr_LV.csv (28.6 KB)
20221207_Heart_MetaboAnalyst_Input_Meta_for_2Way_LV.csv (697 Bytes) - Document all steps leading to the issue. Sometimes screenshots may be necessary
Used Parreto Scaling with no filtering. PCA worked fine, but 2way ANOVA have unknown error. - If it is about using R packages, please provide the environment information, such as sessionInfo()
Not using an env, just using the online MetaboAnalyst website interface.
Any help you can give would be greatly appreciated!
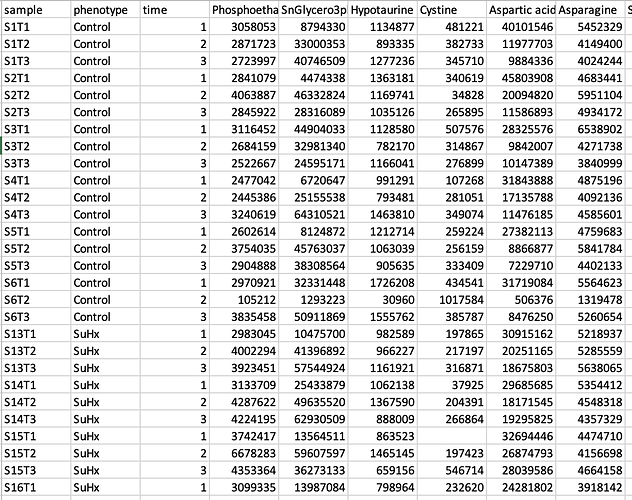
The issue is that you’ve still included the metadata in your concentrations table:
If you look at the example data, when a separate metadata table is uploaded, you should not also include this information in the concentration file. MetaboAnalyst is treating everything except for the first column as concentrations, which breaks the ANOVA module. When I delete the ‘phenotype’ and ‘time’ columns, it’s working.