Hello everyone! I am seeking guidance on troubleshooting my analysis with MetaboAnalyst.
Data Background: I am trying to analyze MRM-profiling lipid data that represent relative quantities of lipid species in an animal’s liver. My study had 5 exposure groups and a control group. Metadata table includes chemical treatment, spatial block, animal developmental stage (NF), and animal sex. I chose to use the linear model with covariate adjustment due to unbalanced groups.
1. Which tool and which module?
* MetaboAnalyst —> Statistical Analysis [metadata table] —> Linear Model with Covariate Adjustment
2. Provide a copy of your data (actual data that leads to the error, not a screenshot image of the data), or indicate which example data you use
DG-Consolidated-Relative-NoFault-102023.csv (69.3 KB)
M3-Consolidated-Relative-NoFault-Metadata-102023-CE_DG.csv (4.6 KB)
3. Document all steps leading to the issue. Sometimes screenshots may be necessary
Upload your data and metadata:
* Data type: concentrations
* Study design: Multiple factors / covariates
* Data format: Samples in columns
* Then uploaded data file and metadata file as .csv
Data Integrity check:
* Passed with 140 (samples) by 41 (compounds) data matrix
* 6 groups detected
* No missing values
Meta-data check:
* Passed with 4 metadata factors detected: Treatment, Block, NF, Sex
* Treatment identified as primary metadata (6 groups)
Normalization Overview:
* Sample normalization: None
* Data transformation: None
* Data scaling: Autoscaling
Metadata visualization:
* Covariates not highly correlated
Univariate Analysis
* Linear Models with Covariate Adjustment
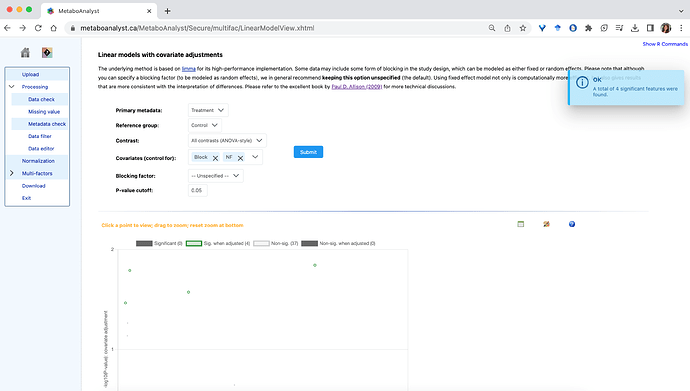
* Started by using the All contrasts (ANOVA-style) using my ‘Block’ and ‘NF’ covariates, which worked:
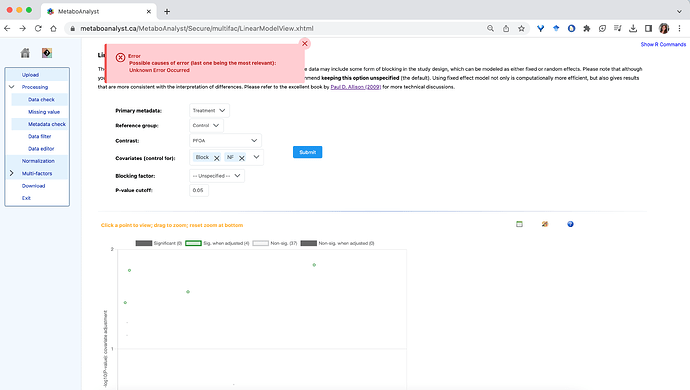
- All individual contrasts can be run, except for the PFOA treatment:
The same error is received when attempting to run this contrast with any individual covariate, or without covariate specified.
Some troubleshooting steps I have attempted after searching the forum and checking guidelines:
- Cleared my internet cache and ensure running one tab on my browser
- Cross compared sample names between the data table and metadata. I could not identify mismatches.
- Checked the number of samples per group (n > 3)
- ‘Find & Replace’ function on excel to replace any rogue space (“ “) detected in the .csv with no space (“”)
- Tried using .txt files instead of .csv files
- Cross compared format with TCE and Covid example data/metadata
- Tried replacing ‘sample’ with ‘#NAME’ in data/metadata
- Created new .csv files with only Control & PFOA data to see if they would run (and they did not work).
This error can not be reproduced using the TCE example data, so I am certain there is something wrong with my data/formatting that I cannot identify.
I have been encountering similar ‘Unknown Errors’ with other dataframes & metadata tables for this project, and have been also unsuccessful identifying formatting issues or other potential causes. Any assistance in troubleshooting this lipid class would be greatly appreciated (so I can then hopefully troubleshoot the others successfully on my own).
Thank you to the Xia lab (and community) for their time and effort in creating/maintaining these tools!