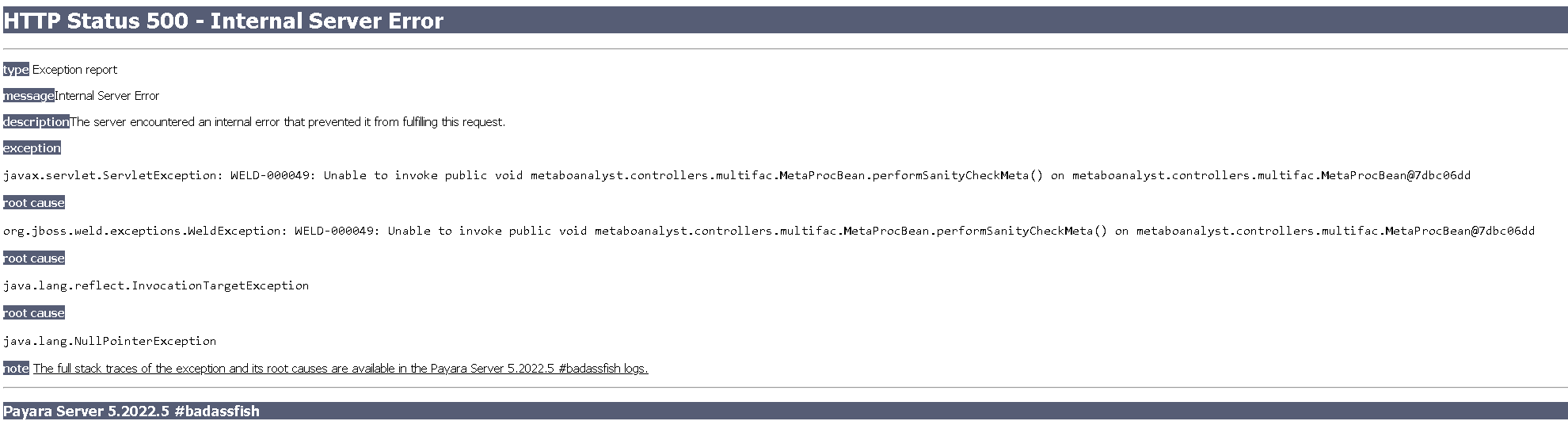
Please help!
I successfully uploaded the data and metadata files (in .csv, mirroring example for TCE data), and when I try to proceed with the Data Integrity check, I reach an error
Attached are the two files.
20230818_SSF metadata_complete_no spaces.csv (6.0 KB)
20230818_SSF_feature list with metadata_multivariate.csv (1.6 MB)
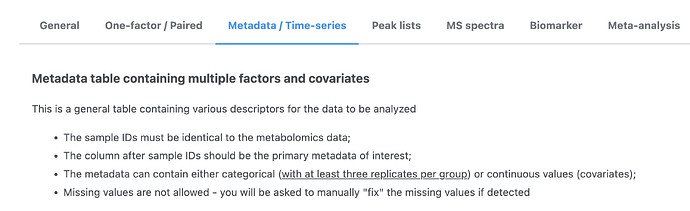
I would strongly suggest to read our the detailed instructions on Data Formats. It gives the clear rules on how to prepare your metadata.
A screenshot for the metadata instruction is shown below
Hi Jeff,
Thanks for your speedy reply.
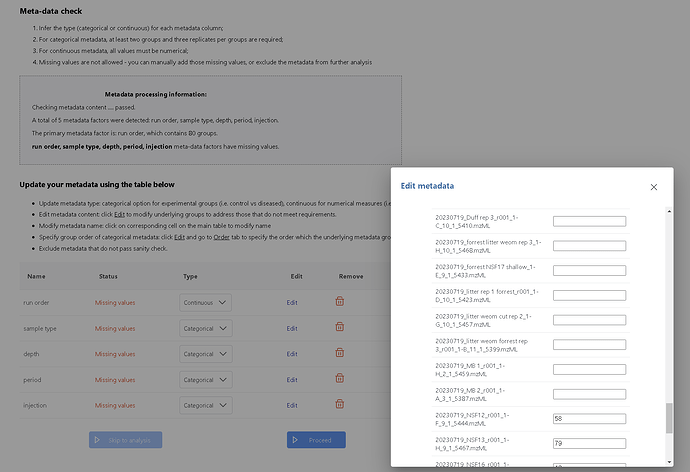
I updated my metadata table to eliminate empty values, however when I submit the file for sanity check the values are not present, see screen shot and modified files
I have changed the “run order” variable to continuous–basically I want to differentiate to see if there are some run order effects.
I also adjusted the metadata to remove categories like “run order” that don’t have replicates, but run into the same issue.
As far as I can tell, the file names all match, except for the metadata to be accepted i had to delete spaces in the metadata (but I did not delete in the filename in the feature list. Could this be a problem?
20230822_SSF metadata_complete_no spaces_filled_reduced variables.csv (5.0 KB)
20230822_SSF_feature list_no metadata_sample header.csv (412.0 KB)
Thanks!
Please perform a quality check on your data - I see so many sample names are inconsistent or missing between the data and the metadata. The issue can only be addressed by the researchers who created such files. In addition, I strongly suggest to use short sample names, as the graphics will become very crowded or cutoff.
The website will be updated in the weekend to give more informative messages. I copy some output if you would like to work on it:
Sample names missing in metadata file: 20230719_CL rep 1_r001_1-C_11_1_5412.mzML; 20230719_cut litter weom rep 3_r001_1-D_9_1_5422.mzML; 20230719_Duff rep 1_r001_1-C_9_1_5409.mzML; 20230719_Duff rep 2_r001_1-F_10_1_5445.mzML; 20230719_Duff rep 3_r001_1-C_10_1_5410.mzML; 20230719_forrest litter weom rep 3_1-H_10_1_5468.mzML; 20230719_forrest NSF17 shallow_1-E_9_1_5433.mzML; 20230719_litter rep 1 forrest_r001_1-D_10_1_5423.mzML; 20230719_litter weom cut rep 2_1-G_10_1_5457.mzML; 20230719_litter weom forrest rep 3_r001_1-B_11_1_5399.mzML; 20230719_MB 1_r001_1-H_2_1_5459.mzML; 20230719_MB 2_r001_1-A_3_1_5387.mzML
Thank you, Jeff.
A problem I ran into while uploading, was that the metadata file would not be accepted if there were spaces in the sample name. To pass the check, I deleted the names of the metadata, but I guess I needed to change the file names as well.
I am using MZmine to pre-process data, and as that software is very sensitive to the raw file name (especially with their metadata settings…which frankly don’t work), our sample names are the same as on acquisition.