I would like to know why compounds with P value>0.05 can be matched into the pathway diagram. Is there any way to filter out these compounds?
For example: In this metabolic pathway with p value = 0.003, the orange compound actually has a p value of 0.07. Can the enrichment pathway analysis then be trusted?
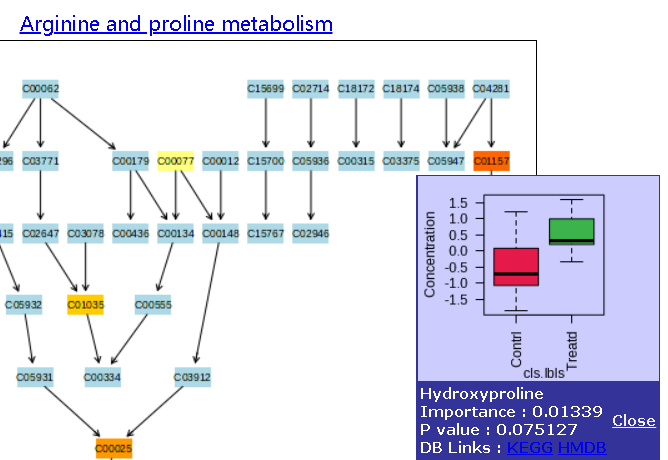
Your question is relevant to over-representation analysis (ORA). I guess you are using globaltest / GlobalANCOVA for analysis in this case, since no details were provided.