MIMMM
March 29, 2023, 6:27am
1
[Raw Data Processing] option in MicrobiomeAnalyst
The error are as follows:
Dereplication failed
Analysis failed at “Step 3: Perform Sequencing data dereplication.”
I have tried uploading new raw data and adjusting the trimming parameters, but the issue still persists.
High number of denoised read
Prior to the above error, there was one instance where the analysis completed up to “Step 8: Perform Sequencing taxonomy assignment.”
However, all samples had a high number of denoised reads, resulting in a very low number of merged reads for analysis.
I would like to inquire about how to adjust the trimming parameters to address this issue.
And, I sent the data related to it to the email address yao.lu5@mail.mcgill.ca.
Thank you very much!
Yao
March 30, 2023, 5:35am
2
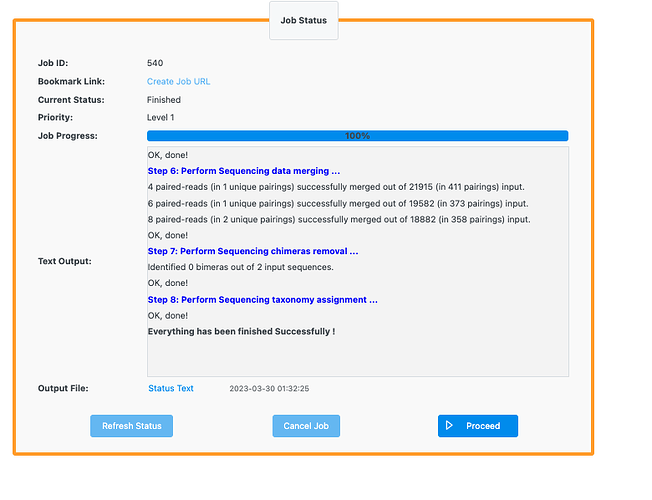
Hello, I have tested your data and it actually works as shown below:
I’m not quite sure about the issue.Could you please try again and let us know if it works? Maybe you can try another browser.
MIMMM
March 30, 2023, 5:54am
3
I have tried it on another browsers and even on another computer, but it still doesn’t work.
Also, may I please request an answer to my second question?
Thank you very much for your help!
Yao
April 2, 2023, 4:13am
4
Hello, for the processing, you can provide us your job id and then we can check the exact issue.
And for your second question, it might be related to how you trim the sequence. You can also refer to:
Hi everyone, I am blocked on my quality filtering part of analysis and I need some help. I used dada2 denoising on my microbiome analysis and it seems there are many reads filtered out on the way. In my opinion too many. I have searched through...
Reading time: 4 mins 🕑
Likes: 17 ❤
I have a question about the counts of number of denoised reads in the denoising-stats of dada2. When I run dada2 in R as shown in the dada2 tutorial (https://benjjneb.github.io/dada2/tutorial.html), different numbers of denoised reads were obtained...
Reading time: 1 mins 🕑
Likes: 11 ❤
for more information.
Hope this helps!
MIMMM
April 2, 2023, 11:09pm
5
Thank you very much!
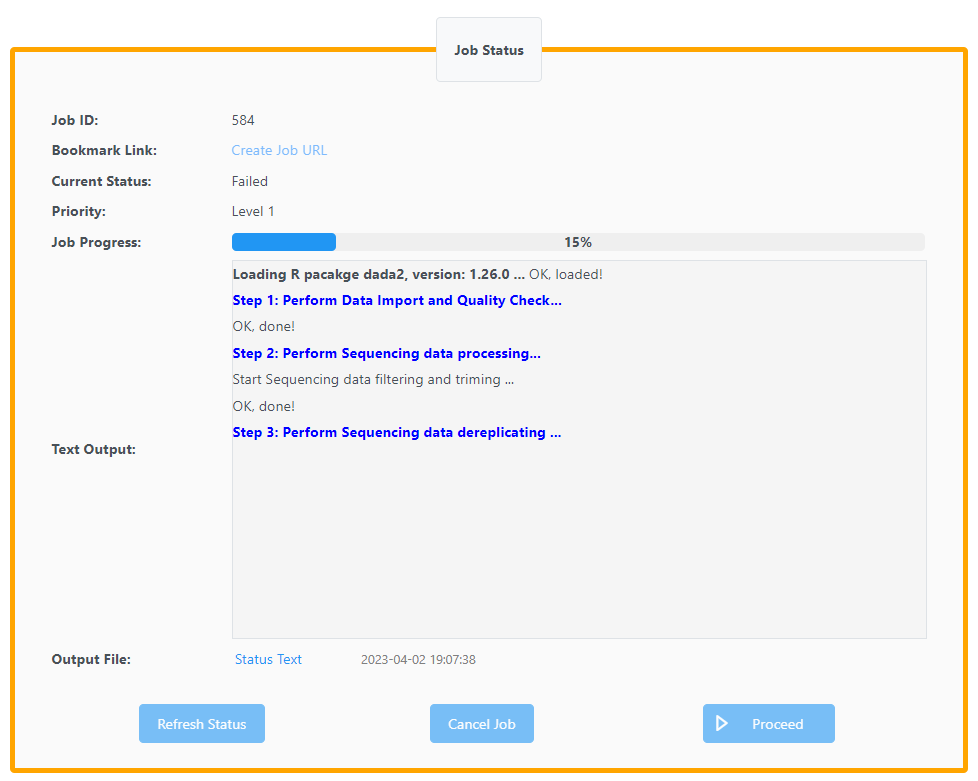
My job id is 584.
Please check the exact issue
Naomi
April 3, 2023, 9:19pm
6
I am having the same issue with preprocessing failing at the data dereplication step. Please let me know if you figure out how to solve this!
Yao
April 4, 2023, 4:39am
7
Hello, the problem was caused by the trim parameters you have set. No reads passed the filter step. You need to modify the parameters.
Yao
April 4, 2023, 4:45am
8
Hello, you can refer to current post or this one to figure out if you have the same problem. If not please provide your job id for further trouble shooting.
xia.lab
April 4, 2023, 11:33am
9
If you have similar issues, please start a new topic and provide Job IDs and parameters settings as the minimum information.