Hello,
I am using MetaboAnalyst website to do a joint-pathway analysis (I am sorry, I am not using R that is why I created a new topic).
I gave a list of genes using entrez ID (I tried with/without fold change) and a list of compounds using KEGG ID (targeted metabolome; I also tried with/without fold change). I am using Drosophila melanogaster organism. When I submitted these lists, I could see a match between all (or almost) entrez ID and genes and between KEGG ID and compounds so the IDs look ok. However, once I clicked on “Proceed” and wanted to visualize my results, I only got results corresponding to the metabolites. For example, if I go to the purine metabolism, I can see in match status “9/148”. All the 9 correspond to a metabolite and none of the gene is red. However, I know some of the genes should be red but for some reason they seem to be not recognized anymore. If I click on “combine p-value (pathway level)” or “gene only”, I got an error saying “Failed to perform integration - not hits found for gene input.”. So the problem is really because of the gene but I do not understand why since after I gave the list, I could see the matches. Do you have any idea what is the problem and what I could do?
Thanks in advance
Please read this post and our response below
Hello,
sorry I will be more specific.
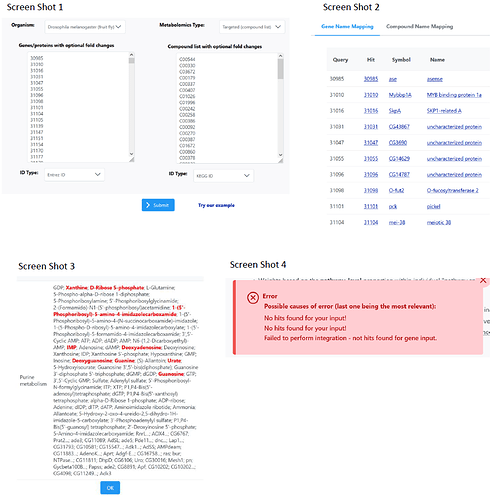
I am using MetaboAnalyst 5.0 (MetaboAnalyst) and then I used the module “Joint-Pathway Analysis”.
As organism, I used “Drosophila melanogaster (fruit fly)” and as Metabolomics type I used “Targeted (compound list)”.
I gave a list a genes with Entrez ID and a list of compound with KEGG ID.
(see screen shot 1):
Then I clicked on “Submit”.
Then I got two tables showing the matches between my genes and compound and the database (see screen shot 2 for genes).
So here everything seems to be fine. So I clicked on “Proceed”.
Then for Pathway Database I chose “Metabolic pathway (integrated)”; for Algorithm Selection I chose “Hypergeometric Test”, “Degree Centrality” and “Combine queries”. Then I clicked on “Submit”.
Then I got some results, however, it seems that no genes are recognized. For example, if I go on “Purine metabolism”, I got a Match Status of 9/148. The 9 are only compounds, no gene appear in red whereas I know that in my list there were AMPdeam and Adk3 (and some others). However they do not appear as being recognized.
(see screen shot 3)
So then, I tried to change parameters (so the step just before seeing results).
For pathway Database, I chose again “Metabolic pathway (integrated)” but for integration method, I chose “Combine p values (pathway level)” and here is the error I got: “Failed to perform integration - not hits found for gene input.”
(see screen shot 4)
That is why I think there is a problem with genes identification but I do not understand why since at the first step, all my genes are recognized but at the end they are not…
Thanks in advance for your help.
Just because a gene is in our annotation database does not mean that it is in the pathway database (but here, it sounds like you have some genes that should be in some pathways). If you send your lists to jessica.ewald@mail.mcgill.ca, I will check to see if this is the case.
Hi Yceyh001,
There was an internal issue for Gene matching. We have fixed it. MetaboAnalyst will be updated in 72h.
Zhiqiang
Hello,
ok great. Thanks to all of you for your help