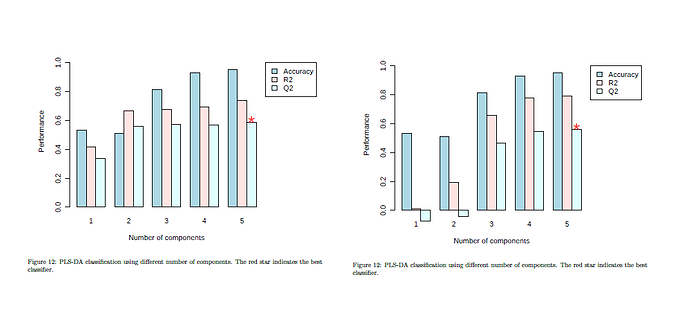
Hello everyone!
While redoing some PLS-DA analyses in MetaboAnalyst, I encountered the problem that the Q2 and R2 values in the cross-validation are highly different when choosing different group labels. Accuracy does not change. Investigating the problem, I found that the alphabetical or numerical order of the group labels does influence this phenome. I’m aware that the PLS-DA is taking the group order into account, but unchecking the respective setting should eliminate that problem. I’m relatively new to this concept so I might miss something. Could anyone clarify this problem?
Thanks in advance!
PLS-DA R2/Q2 are quantitative measures and are more sensitive to model variations (such as random partitions during CV) compared to Accuracy. If you have a small sample size, such variations can be quite noticeable. You can repeat analysis (keep clicking Update button) to see if the result is stable or not.
Thanks for the quick response, Dr. Xia!
In fact, my sample size is relatively small.
However, I think I did not explain my problem clearly: I tried the same sample set several times with different labels: e.g. Control, Hypoxia15 and Hypoxia30 or Normoxia, Hypoxia15 and Hypoxia30. The sets correspond to the first or second plot of my screenshot, respectively. Renaming “Control” to “Normoxia” changes the alphabetical position of this group during processing in MetaboAnalyst, which also can be seen in the downloaded processed data. To my view, this does causes the different CV plots. Using “update”, the respective plots do not change noticeably.
Therefore I do not understand why the group label does influence the outcome even though I unchecked “class order matters” and the group partitions during CV should be random.
OK, here is the paradox you need to solve first:
-
If your group orders are meaningful, you can use regression based measures (R2/Q2), but you should keep group orders intact
-
If your group orders are not meaningful, you can ignore order but should use classification based measure (Accuracy) - maybe we should hide or disable R2/Q2 measures once users uncheck the option
Overall, multi-group classification with small sample sizes is “more of an art”, and is not very useful especially for PLS-DA (it could overfit even for ~100 samples in two group classification!). Discussing performance in this scenario does not carry too much meaning. Random forests is more robust in this case