Hi
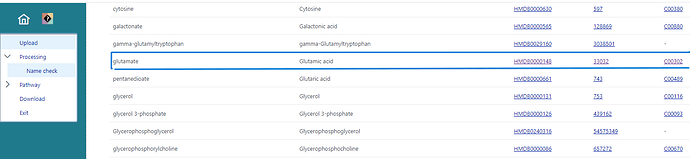
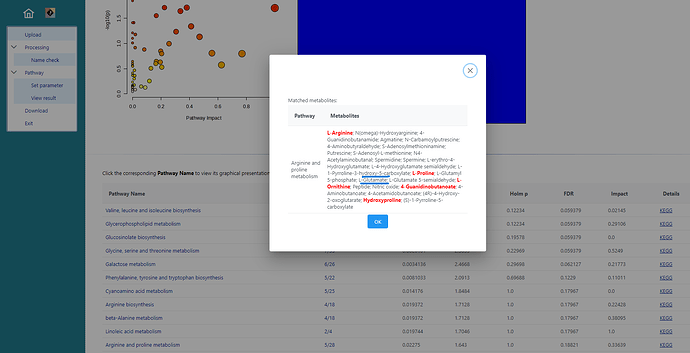
I am using metaboanalyst online server for pathway analysis using Zea myas library and I notice that few important compounds like Glutamate and Alanine are not used/highlighted in particular pathways even after matching the compound names in the library. This is affecting the overview of my metabolomics study since due to missing compounds (glutamate/ alanine), the -log 10 p values and FDRs are going low. Appears to be some technical glitch. It would be helpful if it could be resolved.
Attaching the screenshots from my analysis for your reference.
Thanks