I have a question. In trying to use the pathway analysis, we ran into some issues with the name check. Using KEGG IDs, and the Metaboanalyst ID conversion, the resulting set of annotations were not found when uploading the final data.
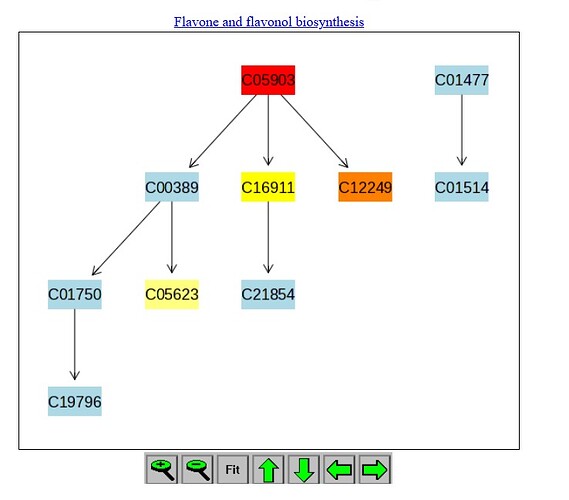
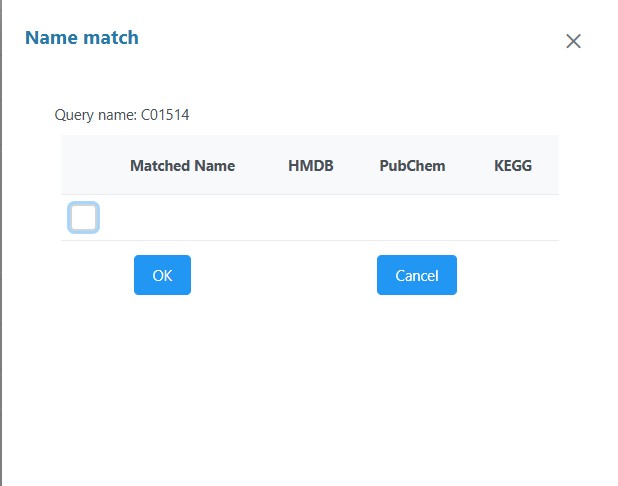
As an example, the data contained “luteolin” which was annotated to C01514. This was a row in the dataset, but when uploading, the name check failed to correlate C01514 to any known metabolite in the KEGG dataset.
Even clicking on the ‘view’ failed to find any IDs
BUT, when running the dataset (after having C01514 omitted), the pathway analysis had a spot for C01514 in the flavone pathway!
Why didn’t the name check find this metabolite, which is common enough to be within the annotated pathways that Metaboanalyst is using to construct the analysis? This was not the one which was identified prior to upload but the name check didn’t ‘find’ a match.
Thank you for your help!