Hello, first time using metaboanalystR
I am looking at changes in metabolites from a cohort of human samples, comparing two groups, those infected with a specific parasite and those without.
i have been following/working through the tutorial from here:
Statistical Analysis
I have normalized in the following way
mSet<-Normalization(mSet, “MedianNorm”, “LogNorm”, “AutoNorm”, ratio=FALSE, ratioNum=20)
The issue seems to be with the P-values so here is the code from the t-test calculations
mSet<-Ttests.Anal(mSet, nonpar=F, threshp=0.05, paired=FALSE, equal.var=TRUE, “fdr”, FALSE) #p-values calculated
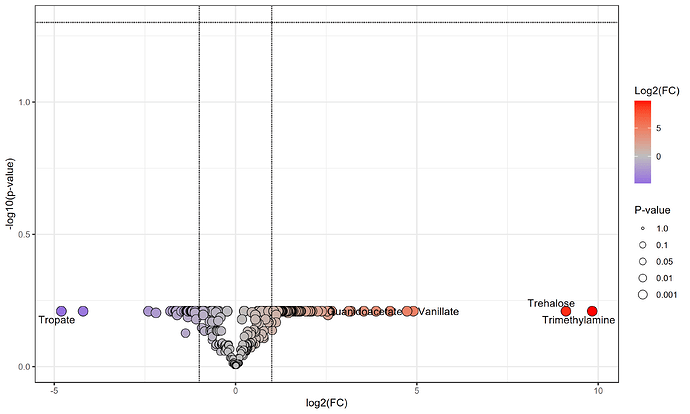
this is the resultant plot
It looks incredibly flattened along the p-value axis. Is this simply because i have no significant metabolites or is it due to something else. Apologies for the potential basic error i am very new to metabolomics
help super appreciated!
-Wedwards