Hello,
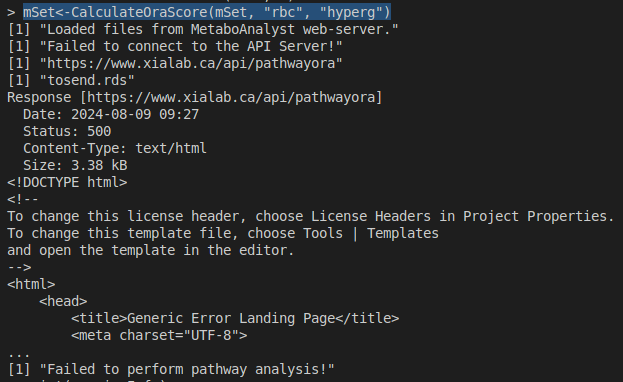
I’ve been working on a Pathway Analysis tutorial locally, but I’m getting a 500 error from the server. Is the server still down, or is there an issue with my tosend.rds file? If the server is still down, what alternative solutions are available?
RDS File:
readRDS(“tosend.rds”)
$mSet
$mSet$dataSet
$mSet$dataSet$type
[1] “conc”$mSet$dataSet$design.type
[1] “regular”$mSet$dataSet$cls.type
[1] “disc”$mSet$dataSet$format
[1] “rowu”$mSet$dataSet$paired
[1] FALSE$mSet$dataSet$pair.checked
[1] FALSE$mSet$dataSet$cmpd
[1] “Acetoacetic acid” “Beta-Alanine” “Creatine”
[4] “Dimethylglycine” “Fumaric acid” “Glycine”
[7] “Homocysteine” “L-Cysteine” “L-Phenylalanine”
[10] “L-Serine” “L-Threonine” “L-Tyrosine”
[13] “L-Valine” “Phenylpyruvic acid” “Propionic acid”
[16] “Pyruvic acid” “Sarcosine”$mSet$dataSet$q.type
[1] “name”$mSet$dataSet$map.table
Query Match HMDB PubChem KEGG
[1,] “Acetoacetic acid” “Acetoacetic acid” “HMDB0000060” “96” “C00164”
[2,] “Beta-Alanine” “beta-Alanine” “HMDB0000056” “239” “C00099”
[3,] “Creatine” “Creatine” “HMDB0000064” “586” “C00300”
[4,] “Dimethylglycine” “Dimethylglycine” “HMDB0000092” “673” “C01026”
[5,] “Fumaric acid” “Fumaric acid” “HMDB0000134” “444972” “C00122”
[6,] “Glycine” “Glycine” “HMDB0000123” “750” “C00037”
[7,] “Homocysteine” “Homocysteine” “HMDB0000742” “91552” “C00155”
[8,] “L-Cysteine” “L-Cysteine” “HMDB0000574” “5862” “C00097”
[9,] “L-Phenylalanine” “Phenylalanine” “HMDB0000159” “6140” “C00079”
[10,] “L-Serine” “Serine” “HMDB0000187” “5951” “C00065”
[11,] “L-Threonine” “L-Threonine” “HMDB0000167” “6288” “C00188”
[12,] “L-Tyrosine” “L-Tyrosine” “HMDB0000158” “6057” “C00082”
[13,] “L-Valine” “L-Valine” “HMDB0000883” “6287” “C00183”
[14,] “Phenylpyruvic acid” “Phenylpyruvic acid” “HMDB0000205” “997” “C00166”
[15,] “Propionic acid” “Propionic acid” “HMDB0000237” “1032” “C00163”
[16,] “Pyruvic acid” “Pyruvic acid” “HMDB0000243” “1060” “C00022”
[17,] “Sarcosine” “Sarcosine” “HMDB0000271” “1088” “C00213”
SMILES Comment
[1,] “CC(=O)CC(O)=O” “1”
[2,] “NCCC(O)=O” “1”
[3,] “CN(CC(O)=O)C(N)=N” “1”
[4,] “CN(C)CC(O)=O” “1”
[5,] “OC(=O)\C=C\C(O)=O” “1”
[6,] “NCC(O)=O” “1”
[7,] “NC@@HC(O)=O” “1”
[8,] “NC@@HC(O)=O” “1”
[9,] “NC@@HC(O)=O” “1”
[10,] “NC@@HC(O)=O” “1”
[11,] “CC@@HC@HC(O)=O” “1”
[12,] “NC@@HC(O)=O” “1”
[13,] “CC(C)C@HC(O)=O” “1”
[14,] “OC(=O)C(=O)CC1=CC=CC=C1” “1”
[15,] “CCC(O)=O” “1”
[16,] “CC(=O)C(O)=O” “1”
[17,] “CNCC(O)=O” “1”$mSet$dataSet$use.metabo.filter
[1] FALSE$mSet$analSet
$mSet$analSet$type
[1] “pathora”$mSet$imgSet
$mSet$imgSet$margin.config
$mSet$imgSet$margin.config$l
[1] 50$mSet$imgSet$margin.config$r
[1] 50$mSet$imgSet$margin.config$b
[1] 20$mSet$imgSet$margin.config$t
[1] 20$mSet$imgSet$margin.config$pad
[1] 0.5$mSet$msgSet
$mSet$msgSet$msg.vec
character(0)$mSet$msgSet$nmcheck.msg
[1] “1”
[2] “Name matching OK, please inspect (and manual correct) the results then proceed.”$mSet$msgSet$lib.msg
[1] “Your selected pathway library code is \textbf{ hsa }(KEGG organisms abbreviation).”$mSet$cmdSet
character(0)$mSet$paramSet
$mSet$paramSet$report.format
[1] “html”$mSet$paramSet$lib.nm
[1] “hsa”$mSet$return.cols
[1] TRUE TRUE FALSE TRUE FALSE$mSet$lipid.feats
[1] FALSE$mSet$name.map
$mSet$name.map$query.vec
[1] “Acetoacetic acid” “Beta-Alanine” “Creatine”
[4] “Dimethylglycine” “Fumaric acid” “Glycine”
[7] “Homocysteine” “L-Cysteine” “L-Phenylalanine”
[10] “L-Serine” “L-Threonine” “L-Tyrosine”
[13] “L-Valine” “Phenylpyruvic acid” “Propionic acid”
[16] “Pyruvic acid” “Sarcosine”$mSet$name.map$hit.inx
[1] 42 40 46 62 88 78 577 440 103 119 108 102 685 130 158 163 184$mSet$name.map$hit.values
[1] “Acetoacetic acid” “beta-Alanine” “Creatine”
[4] “Dimethylglycine” “Fumaric acid” “Glycine”
[7] “Homocysteine” “L-Cysteine” “Phenylalanine”
[10] “Serine” “L-Threonine” “L-Tyrosine”
[13] “L-Valine” “Phenylpyruvic acid” “Propionic acid”
[16] “Pyruvic acid” “Sarcosine”$mSet$name.map$match.state
[1] 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1$mSet$api
$mSet$api$libVersion
[1] “current”$mSet$api$libNm
[1] “hsa”$mSet$api$filter
[1] FALSE$mSet$api$oraVec
[1] “C00164” “C00099” “C00300” “C01026” “C00122” “C00037” “C00155” “C00097”
[9] “C00079” “C00065” “C00188” “C00082” “C00183” “C00166” “C00163” “C00022”
[17] “C00213”$mSet$api$method
[1] “hyperg”$mSet$api$nodeImp
[1] “rbc”$mSet$pathwaylibtype
[1] “KEGG”$libVersion
[1] “current”$libNm
[1] “hsa”$filter
[1] FALSE$nodeImp
[1] “rbc”$method
[1] “hyperg”$oraVec
[1] “C00164” “C00099” “C00300” “C01026” “C00122” “C00037” “C00155” “C00097”
[9] “C00079” “C00065” “C00188” “C00082” “C00183” “C00166” “C00163” “C00022”
[17] “C00213”
SessionInfo:
sessionInfo()
R version 4.3.2 (2023-10-31)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTSMatrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so; LAPACK version 3.10.0locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=Ctime zone: Etc/UTC
tzcode source: system (glibc)attached base packages:
[1] stats graphics grDevices utils datasets methods baseother attached packages:
[1] MetaboAnalystR_4.0.0loaded via a namespace (and not attached):
[1] DBI_1.2.2 bitops_1.0-7 pROC_1.18.5
[4] rlang_1.1.3 magrittr_2.0.3 scrime_1.3.5
[7] compiler_4.3.2 RSQLite_2.3.5 vctrs_0.6.5
[10] reshape2_1.4.4 stringr_1.5.1 pkgconfig_2.0.3
[13] fastmap_1.1.1 caTools_1.18.2 utf8_1.2.4
[16] prodlim_2023.08.28 purrr_1.0.2 bit_4.0.5
[19] cachem_1.0.8 jsonlite_1.8.8 recipes_1.0.10
[22] blob_1.2.4 BiocParallel_1.36.0 parallel_4.3.2
[25] R6_2.5.1 stringi_1.8.3 RColorBrewer_1.1-3
[28] qs_0.25.7 limma_3.58.1 parallelly_1.37.0
[31] rpart_4.1.21 lubridate_1.9.3 Rcpp_1.0.12
[34] iterators_1.0.14 future.apply_1.11.1 Matrix_1.6-1.1
[37] splines_4.3.2 nnet_7.3-19 igraph_2.0.2
[40] timechange_0.3.0 tidyselect_1.2.0 stringfish_0.16.0
[43] siggenes_1.76.0 timeDate_4032.109 gplots_3.1.3.1
[46] codetools_0.2-19 listenv_0.9.1 lattice_0.21-9
[49] tibble_3.2.1 plyr_1.8.9 Biobase_2.62.0
[52] withr_3.0.0 future_1.33.1 survival_3.5-7
[55] RcppParallel_5.1.7 pillar_1.9.0 KernSmooth_2.23-22
[58] foreach_1.5.2 stats4_4.3.2 plotly_4.10.4
[61] generics_0.1.3 ggplot2_3.5.0 munsell_0.5.0
[64] scales_1.3.0 RApiSerialize_0.1.2 gtools_3.9.5
[67] globals_0.16.2 class_7.3-22 glue_1.7.0
[70] lazyeval_0.2.2 tools_4.3.2 data.table_1.15.0
[73] ModelMetrics_1.2.2.2 fgsea_1.28.0 gower_1.0.1
[76] locfit_1.5-9.8 fastmatch_1.1-4 cowplot_1.1.3
[79] grid_4.3.2 Cairo_1.6-2 impute_1.76.0
[82] tidyr_1.3.1 ipred_0.9-14 edgeR_4.0.16
[85] colorspace_2.1-0 nlme_3.1-163 crmn_0.0.21
[88] cli_3.6.2 fansi_1.0.6 viridisLite_0.4.2
[91] lava_1.7.3 dplyr_1.1.4 glasso_1.11
[94] pcaMethods_1.94.0 gtable_0.3.4 digest_0.6.34
[97] BiocGenerics_0.48.1 caret_6.0-94 htmlwidgets_1.6.4
[100] htmltools_0.5.7 memoise_2.0.1 multtest_2.58.0
[103] lifecycle_1.0.4 httr_1.4.7 hardhat_1.3.1
[106] statmod_1.5.0 bit64_4.0.5 MASS_7.3-60
Any help would be appreciated. Thanks in advance!
All the best,
Gokhan