Hello,
I am trying to recreate MetaboAnalyst’s PLS-DA scores and loadings plots with Python Scikit-Learning PLSRegression functions. For the many different tutorials I have found online, they tend to only give examples for data with only two groups, where they describe assigning the samples in each group either “0” or “1”/“1” or “-1” as a dummy variable for the PLS analysis.
However, my data has many sample groups. This all runs very well in MetaboAnalyst but I am unable to replicate these results myself. Currently, I will assign the samples in the groups “0”, “1”, “2”… “8”, “9” for the dummy variable, but I do not know if this is the correct approach. Also, I’ve seen some tutorials add more columns into the data that assign “1” to the samples in the group of interest and “0” to the others (see image as an example).
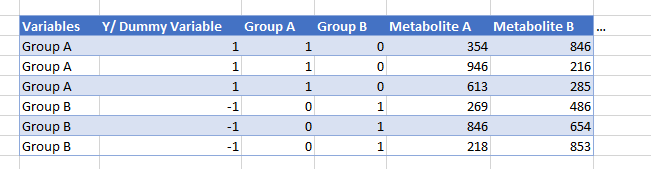
How does MetaboAnalyst process the normalized data in its PLS-DA pipeline?
Any help would be much appreciated.
Many thanks,
Jess