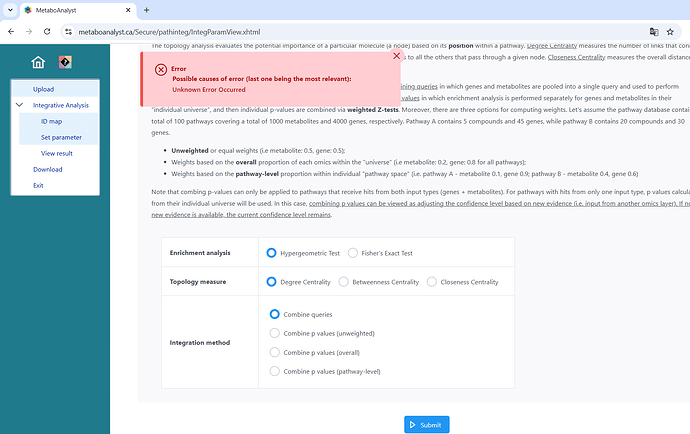
I had done a joint pathway analysis on saccharomyces cerevisiae a few days ago and it works normally.
Now, after the update, if I repeat the analysis by inserting the same genes and the same compounds but using the new module it gives me an error, and no gene is found.
| Gene | Fold changes |
|---|---|
| YGL028C | 2.638198017 |
| YOR009W | 2.631340503 |
| YOR247W | 2.459831008 |
| YDR012W | 2.457889275 |
| YOR388C | 2.367131573 |
| YLR445W | 2.312743018 |
| YGR107W | 2.297821834 |
| YNL066W | 2.297637474 |
| YCR034W | 2.296647743 |
| Compound | Fold changes |
|---|---|
| 3-Butyn-1-al | -4.69 |
| Propanoate | -1.16 |
| Glycine | -1.3 |
| Pyrimidine | -1.3 |
| 5-Aminoimidazole | 2.59 |
| Piperidine | -0.66 |
| Crotonate | -2 |
| 2-Aminoprop-2-enoate | -1.08 |
| D-Alanine | -1.17 |
I used gene symbols and compound names as the input, and selected Saccharomyces cerevisiae as the organism in the new module. The error is “no gene hits” but before the update it found them.