Hello,
I am experiencing issues when processing my LC-MS/MS data in MetaboAnalyst 6.0, specifically in the LC1+DDA
Experimental Setup
- Instrument: Q Exactive (Thermo Scientific)
- Acquisition Mode: DDA
- Ionization: Both positive and negative modes in the same run
- MS2 Parameters:
- Resolution: 17,500
- AGC Target: 1e5
- Loop Count: 5
- Isolation Window: 1 m/z
- NCE: 20, 30, 40
- Profile Mode (not centroided)
Data Processing Steps Taken:
-
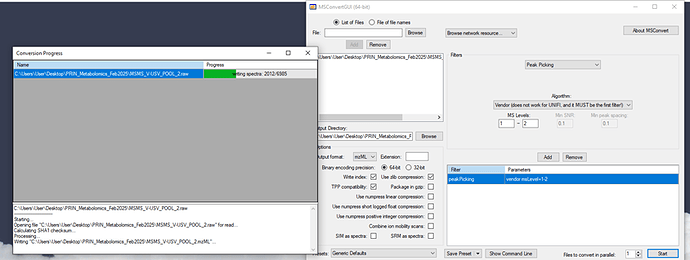
Raw data were converted to mzML format using MSConvert, following the guidelines in the MetaboAnalyst 6.0 tutorial.
-
Metadata and sample files were properly prepared.
-
The dataset was uploaded using the MS1+DDA mode.
-
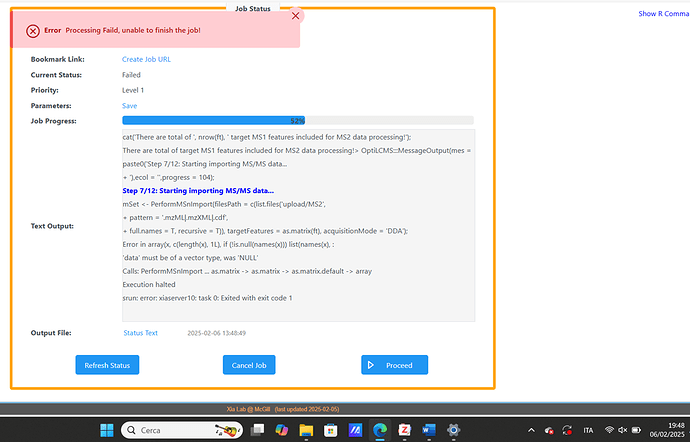
However, I encountered an error during the MSMS processing step.
I was wondering if the fact that my data were acquired in profile mode (not centroided) could be causing this issue. Should I reprocess my data in centroid mode before conversion?
Further Testing: LC-MS1 Only Analysis
Since the MSMS processing failed, I also tried to process the data using only LC-MS1 mode.
- Unfortunately, the results were no features identified.
- I tested all available Peak Picking Algorithms, but the results remained the same.
- Additionally, there was almost no match with HMDB, which seems unusual.
- Here a screeshot from the generated report
Questions:
- Could the profile mode acquisition be causing issues with MetaboAnalyst’s processing?
- Are there specific settings or preprocessing steps I should adjust to improve feature identification?
- Has anyone experienced similar issues when using DDA, or when trying to process MS1-only data?
Any help or insights would be greatly appreciated!
Thank you in advance.
Best,
Giulia