I am trying to run a beta diversity test when using a .biom file, metadata file, and a .tre phylogenetic file (with GreenGenes Taxonomy labels). I am uploading data in the BIOM format, and have separate files for metadata, .biom, .tre phylogenetic file, and am putting in GreenGenes Taxonomy as the Taxonomy labels.
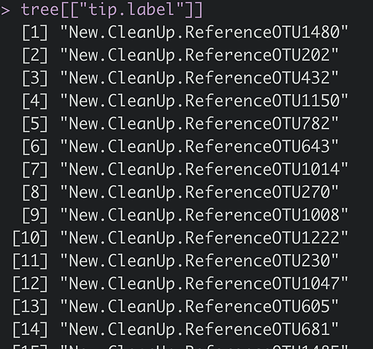
In my distance method I am using an unweighted UniFrac distance, and I am getting the error: “Tree tip labels do not match feature names in the OTU/taxonomic tables!”. Other tests that don’t rely on the phylogenetic file work (like using other distance methods such as Jensen-Shannon Divergence). What could possibly be causing the issue?
I am using the .tre phylogenetic file from here: Importing data — QIIME 2 2022.8.3 documentation (for some reason I can’t upload it here), is it possible that the GreenGenes taxonomy file isn’t compatible with the unrooted-tree.tre file? A link to the exact data file I am inputting is here: unrooted-tree.tre - Google Drive.