A nice introduction to logistic regression (LR) can be found here. To prepare the input data, it is recommended to use the number (0 or 1) instead of string labels. Usually, 1 is used for the case and 0 is for the control.
LR usually works best for a dozen or so variables and cannot be directly used for omics scale data with 100s ~1000s of variables. It is often necessary to perform feature selection first. MetaboAnalyst offers this support in its Biomarker Module under the “ROC curve based model evaluation (Tester)” track, since this track allows users to manually select a subset of features/samples for ROC analysis using the statistics table (feature ranking) in the Model Builder step
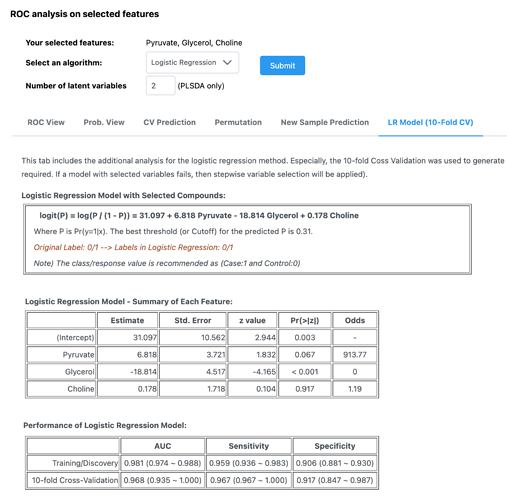
You can then perform logistic regression analysis with the selected variables and explore the modelling results. The results include plots and tables are generated using MCCV (100-fold cross-validation). In addition, it produces the result with 10-fold cross-validation in the “LR model (10-fold CV)” tab. An example output is shown below:
Cautions! if you have selected features based on their overall ranks (AUC, T-statistic or fold changes), there will be an increase risk of overfitting - they may be the best biomarkers for this particular data, but not the case for new samples!