Many normalization and differential expression analysis methods do not perform well when there are missing entries in the data matrix. One option is to filter out genes with too many missing values. Another is to guess, or impute, reasonable values for the missing values. In EcoToxXplorer, we assume that missing values represent situations where there is very low or no expression, and so we replace them with a high Ct value. Replacing all missing measurements the exact same value causes problems during the statistical analysis, potentially leading to artificially low p-values due to the low variance.
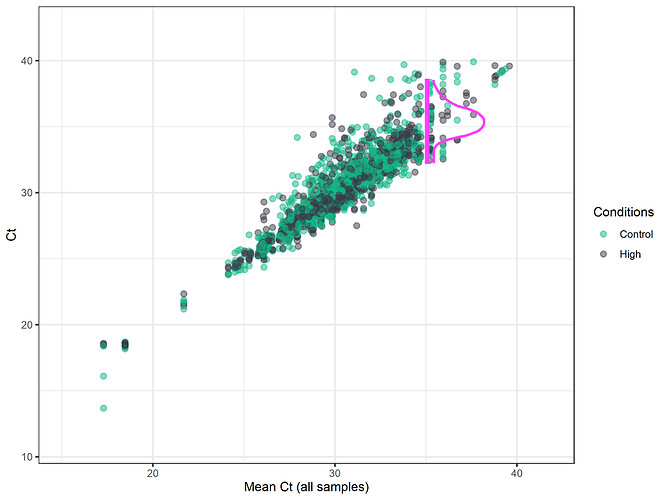
The first step in EcoToxXplorer imputation is to decide the Ct cut-off for reliable measurements, as accuracy tends to decrease at high cycle numbers. Then, all missing values and values above the cut-off are replaced by randomly drawn values from a normal distribution that has a mean of the Ct cut-off and a standard deviation of the original data surrounding this cut-off. For example, in the image above, the Ct-cutoff was chosen to remain at the default of 35. Then, values are randomly drawn from the distribution of the data around this point.
You can always turn off imputation by unselecting the “Impute non-detects” check box on the “Data Normalization” page, in which case genes with less than 3 observations per experimental group will be filtered out. You can view a summary of how many missing values were imputed for each experimental group by viewing the “Processing Summary” tab.