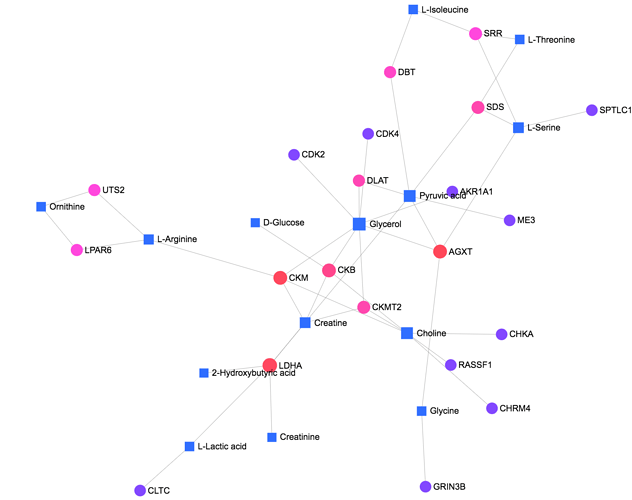
The networks are generated by first mapping the metabolites, genes or both to the underlying gene-metabolite-disease interactions database.
A search algorithm is then performed to identify first-order neighbours (e.g. metabolites that directly interact with a given metabolite) for each of these mapped metabolites (“seeds”). The resulting nodes and their interaction partners are returned to build the subnetworks.
The above figure shows an example of a generated metabolites-genes interaction network.