Hi,
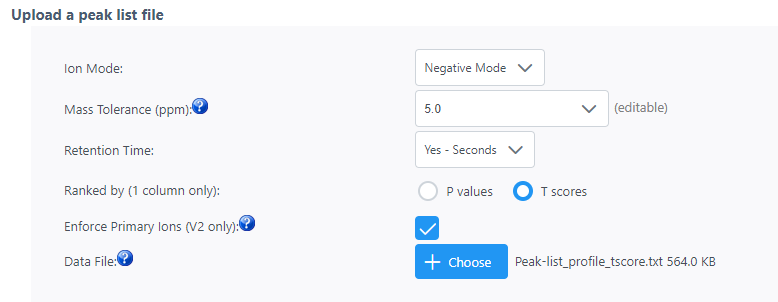
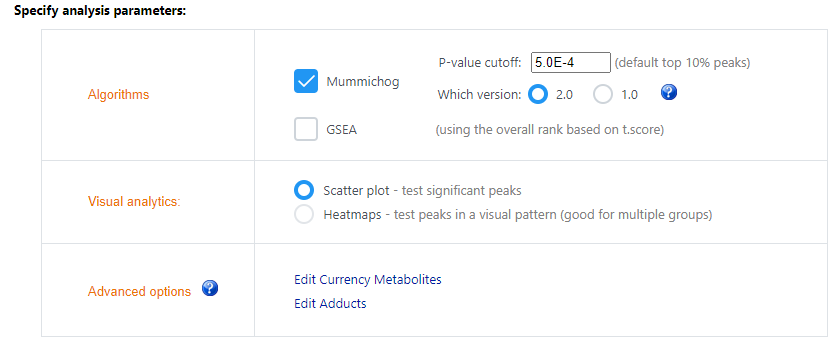
I am currently using your Functional Analysis module to predict changes in metabolic pathways between two sample conditions after LC-MS analysis. I uploaded a peak list file with t-scores (see attachments) and chose the mummichog algorithm, but I would like to have some precision about the results that I have attached.
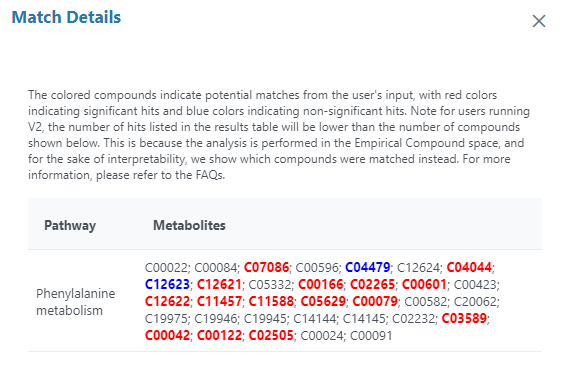
Indeed, for the phelalanine metabolism pathway for example, 45 hits are indicated in the result table, with 25 significant hits among them. But when I open the match details, I see in total 33 compound IDs, 2 of them being colored in blue, and 15 of them colored in red. It is indicated in the match details explanation that red compounds are significant hits, so I wondered why there are only 15 of them, and what do the non-colored compounds represent ? If blue compounds are the non significant ones, shouldn’t we see 20 of them ?
Also, what is the difference between a “match” and a “hit” ?
Thank you in advance for your help.