Dear Dr. Xia,
I encountered a few errors in the LC/MS Spectral processing tool of Metaboanalyst v6.0. I have uploaded a demo video showing the errors and a PPT mentioning the errors in detail for your perusal.
Some of the errors highlighted are : 1) Not supporting metadata file, 2) Blank subtraction option not functioning, 3) Not able to understand the data is in centroid mode, etc.
Please accept our thanks in advance. Looking forward to your positive response.
The link of the files - Dropbox
Even you did not provide actual data and study design (this is a general request for troubleshooting), the issues are clear based on your slides:
-
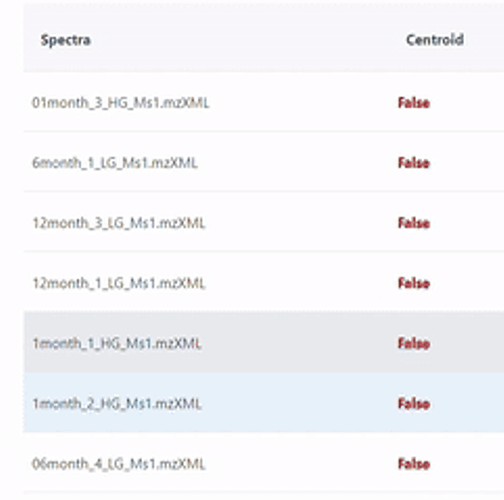
Spectra data MUST be in centroid mode - it seems not the case - your slides show that all spectra are NOT in centroid mode. Note, I also don’t know the instrument type, as you did not mention anything about your data
-
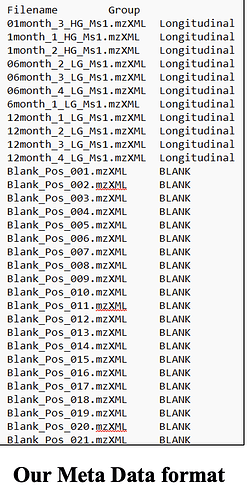
Metadata - there is only one group (exclude BLANK) - this is not permitted. At least two groups with 3 replicated by default.
-
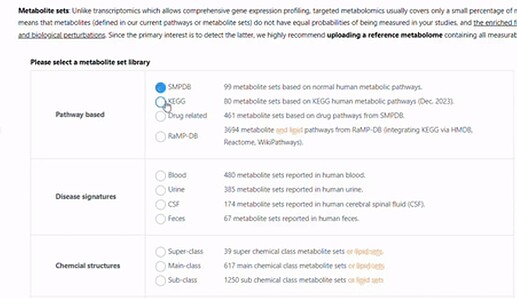
Lipid pathway enrichment analysis - the error message tells you there is no hits in the selected library. This is a fact. Why don’t you choose the libraries containing lipids (see below, their names contains lipids highlighted in yellow).
Finally, please read our tutorials first, rather than uploading data and expect they will work. We also have a week-long or 3-month long training course for omics data analysis