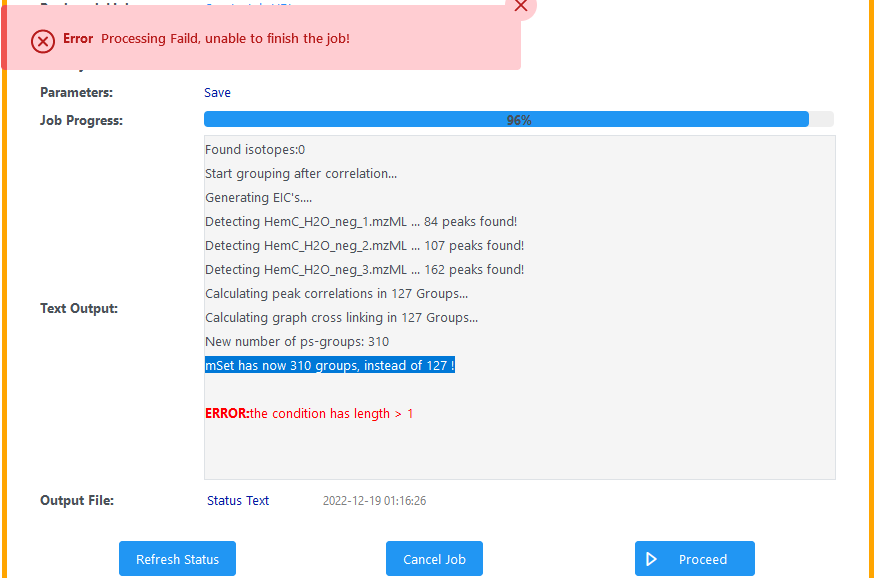
I am processing a raw LC-MS/MS spectra and the analysis halts at the following “mSet has now 310 groups, instead of 127 !” and gives the following error statement, “ERROR:the condition has length > 1”. The data was acquired in UHPLC-QTRAP in negative mode. Can anyone help me out with this?
Hi, please share your job ID or an reproducible example data + parameters with us for further checking.
Hi, here is an example of a job recently submitted (1-2 days ago) which failed in a similar way.
https://www.metaboanalyst.ca/MetaboAnalyst/faces/Share?ID=fwa2rl4it_16880
Since about 1-2 months now, I am unfortunately unable to get any result - even when using data sets which previously worked fine… ![]()
Hi,
The job ID is 16405 and the URL is MetaboAnalyst. I had kept the parameters default as they were.
Hi
Here is another example, failed today, with the same error being returned…
https://www.metaboanalyst.ca/MetaboAnalyst/faces/Share?ID=kekk1mrld_17333
Unfortunately, we are not able anymore to get results from MetaboAnalyst…
Regards
Hello I am getting same problem. Were you able to fix it?
Hi,
Not yet, unfortunately. We are trying to switch to another software for peak picking, and then upload peak list to MetaboAnalyst for further analysis. We are also trying to run the analysis locally using the R package, but so far we are still stuck - for 3 months now
I removed the blank and skipped the blank correction. Now the job was successful. Maybe it is not the ideal setting. Hope it helps!
I tried again, with and without blank correction and with and without contaminants removal; still the same type of error such as:
mSet has now 6898 groups, instead of 754 !
@Qiang @jeff.xia could you please have one more look? A few additional examples:
- MetaboAnalyst → failed at 96%
- MetaboAnalyst → failed at 96%
- MetaboAnalyst → failed at 96%
- MetaboAnalyst → failed at 96%