I have a problem like this while using the Spectra Processing for LC-MS. I use LC-MS/QTOF (HPLC-QTOF) for the platform and use centWave-manual algorithm as the centWave-auto always got me to troubleshooting. I didn’t change anything in the parameters of the centWave-manual.
For the raw data, it is from serum sample from DKD patient and analyze by LC-MS/QTOF Agilent 6510 and the format for the data result is (.d). After that, I transform that into (.mzML) by proteowizard.
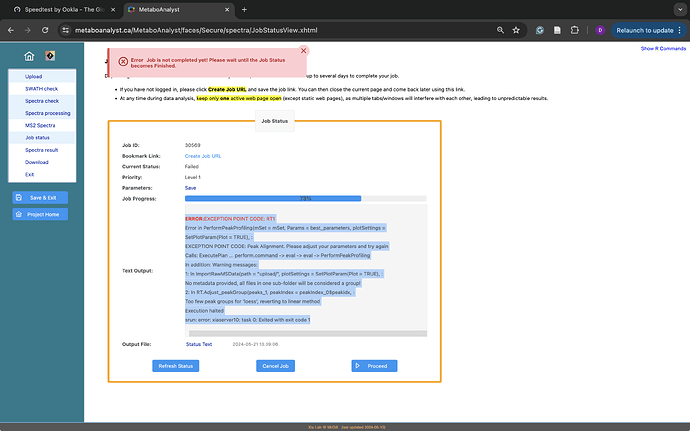
ERROR:EXCEPTION POINT CODE: RT1
Error in PerformPeakProfiling(mSet = mSet, Params = best_parameters, plotSettings =
SetPlotParam (Plot = TRUE),
EXCEPTION POINT CODE: Peak Alignment. Please adjust your parameters and try again
Calls: ExecutePlan … perform.command → eval → eval → PerformPeakProfiling
In addition: Warning messages:
1: In ImportRawMSData(path = “upload/”, plotSettings = SetPlotParam(Plot = TRUE), :
No metadata provided, all files in one sub-folder will be considered a group!
2: In RT.Adjust_peakGroup(peaks_1, peakIndex = peakIndex_0$peakidx, :
Too few peak groups for ‘loess’, reverting to linear method Execution halted
srun: error: xiaserver10: task 0: Exited with exit code 1
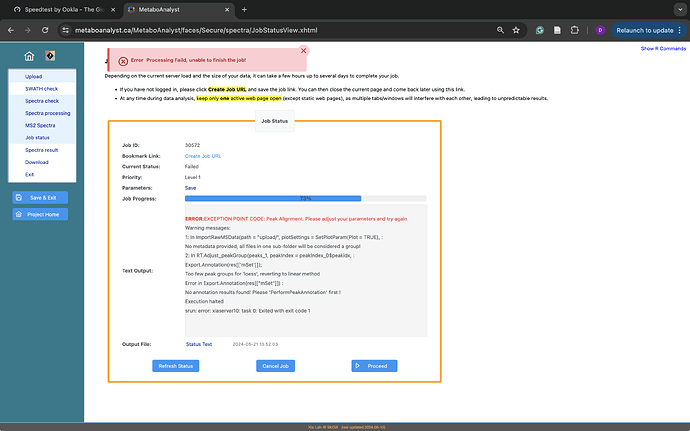
ERROR:EXCEPTION POINT CODE: Peak Alignment. Please adjust your parameters and try again
Warning messages:
1: In ImportRawMSData(path = “upload/”, plotSettings = SetPlotParam(Plot = TRUE), :
No metadata provided, all files in one sub-folder will be considered a group!
2: In RT.Adjust_peakGroup(peaks_1, peakIndex = peakIndex_0$peakidx, :
Export. Annotation(res [[‘mSet’]]);
Too few peak groups for ‘loess’, reverting to linear method Error in Export. Annotation(res[[“mSet”]]) :
No annotation results found! Please ‘PerformPeakAnnotation’ first !
Execution halted
srun: error: xiaserver10: task 0: Exited with exit code 1
Data: SERUM - Google Drive