Hello,
I am analysing RNAseq data on ExpressAnalyst but I cannot perform differential analysis. I upload my file, normalize and filter it, then ask for differential analysis between two groups with Limma statistical method. But then, I have an error message (see attached picture) and the analysis is never completed.
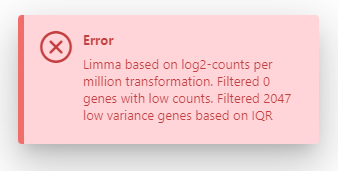
I do not understand because I was doing similar analyses few weeks ago without any issue. Any idea what could be wrong?
Thanks a lot for your help !!!
Hello,
Can you share with us the dataset?
Guangyan
Hi, here is the dataset. I already used this same dataset on ExpressAnalyst and it worked perfectly, but if I remember correctly it was on the previous version where you had to include the metadata in the data file (it was not possible to upload two separate files)…
Dataset.txt (3.2 MB)
Also facing similar problem
Can you please also provide the organism and ID type?
Sure.
Organism: mouse
ID: ensembl gene ID
Thanks for your help!
Hello!
I have the same problem since some weeks ago. I noticed you changed the interface with option for separate metadata file and ever since I can only go till the normalization step, which works as expected. But then in DE I always get a similar error whatever I select and regardless of normalization mode.
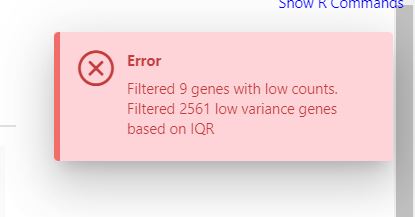
It is interesting that this is the same message I get in the normalization
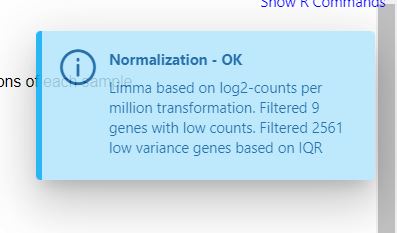
But in that case it’s not an error, but just informative.
Thanks in advance!
Hello,
I have checked your data, the problem was caused by the “+” in your metadata. It will be affected during the processing of Limma. You can manually changed it and try out again for now. We will automatically change “+” to “_” then.
Hope this helps!
You can refer to the solution for [dupuycha] above.
If it is not the same case with your data, please follow our post guideline for detail trouble shooting.
Perfect, it works !! Thank you so much !!
Thanx for the replies! In the meantime I found out that I had some hyphens ( - ) in the names and/or classes of the samples. When I changed them to underscores ( _ ) everything went fine. I had also removed spaces, used only underscores to connect words and also changed all empty cells in the count table to 0s. Not sure if the 0s were needed or if spaces between words can also be accepted - didn’t try each change separately yet - but for sure hyphens are not welcome!
This topic was automatically closed after 11 hours. New replies are no longer allowed.