Hi,
When I use the same metabolites dataset on the Quantitative Enrichment Analysis and Pathway Analysis (human KEGG library used), I am getting different results for the global test. Is this expected?
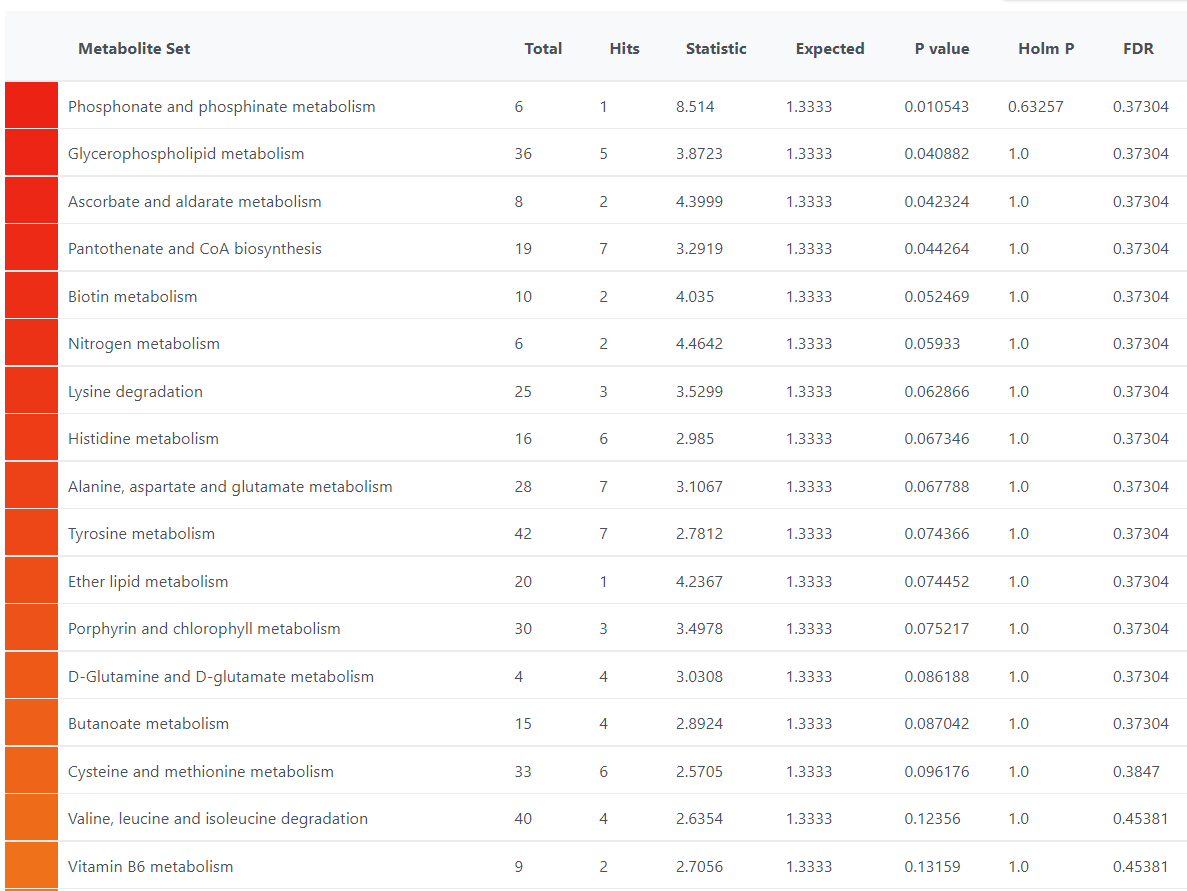
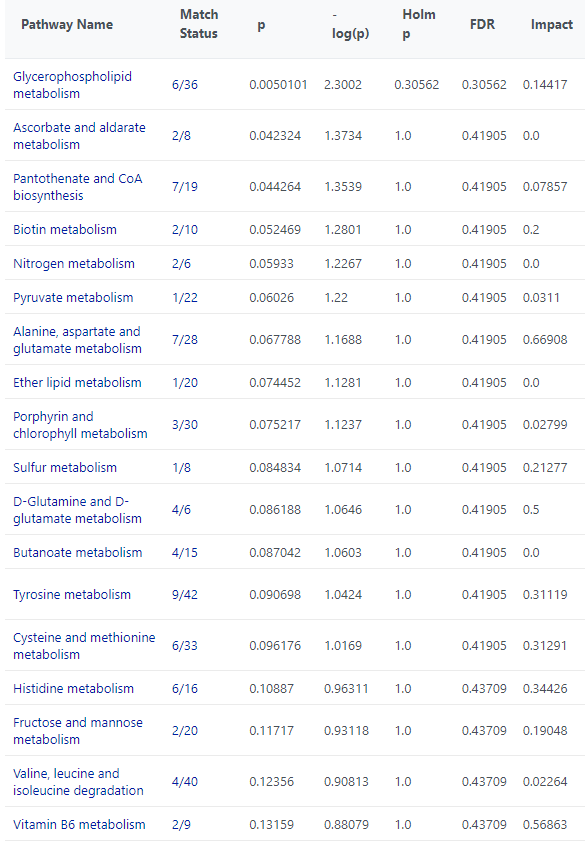
I have noticed that there are differences in the metabolite sets for KEGG used in QEA and Pathway Analysis. For example, the KEGG library used in QEA does not appear to have “Pyruvate metabolism”. But even for the metabolite set that appear the same on QEA and Pathway Analysis, there are difference in the match status.
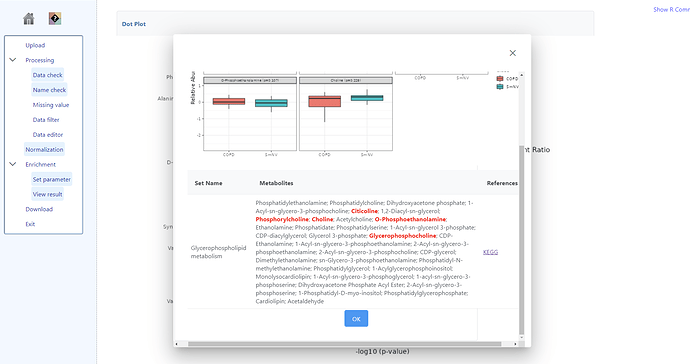
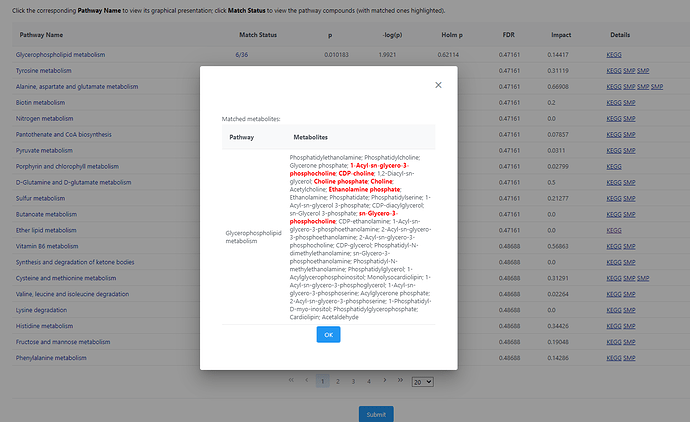
For example, for Glycerophospholipid metabolism there are 5/36 match in the QEA but there are 6/36 match in the pathway analysis. 1-Acyl-sn-glycero-3-phosphocholine was not picked up in QEA.
Which global test result should be used for the enrichment analysis (from QEA module or Pathway Analysis module)?
Thank you
Ken