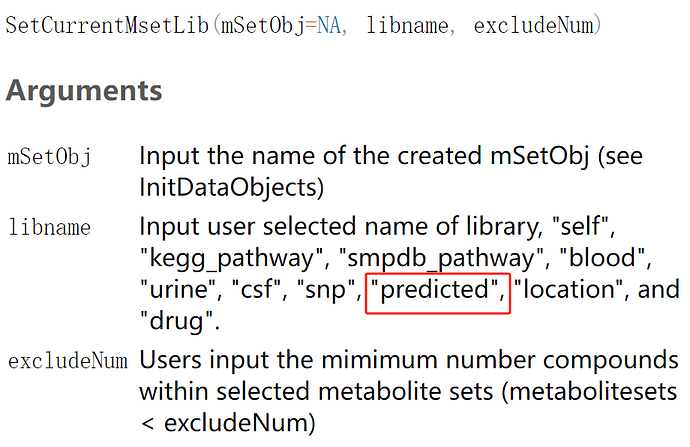
In enrichment analysis using the R function “enrichment analysis”, what are the actual database names corresponding to the libnames “kegg_pathway”, “smpdb_pathway”, “blood”, “urine”, “csf”, “snp”, and “predicted”?
All of our data are documented. I would suggest you to try our web interface to see the results. You can also look inside the database itself to see the details and URLs
This topic was automatically closed after 5 days. New replies are no longer allowed.