Dear All,
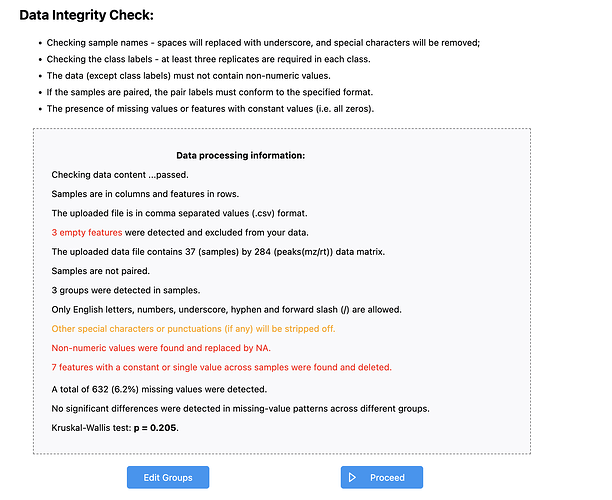
I uploaded my processed proteomics using Generic Format
(.csv)
Statistical Analysis
[one factor]. then using peak intensities, samples unpaired in columns
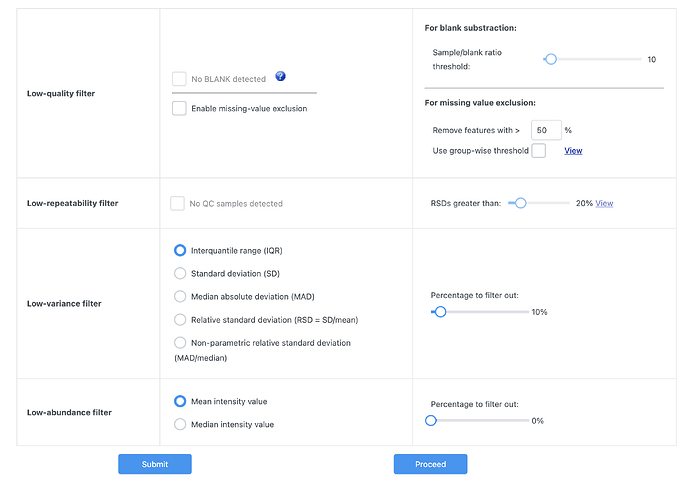
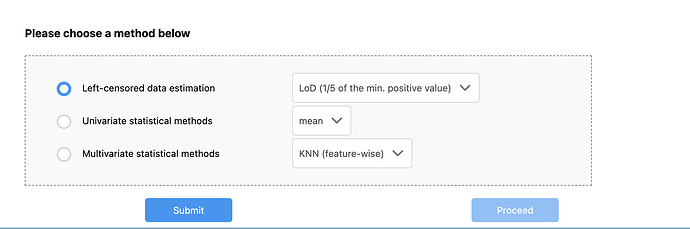
(LC/MS) excel format in the software however I’m facing a problem that I cannot proceed due to missing values in my data set file. the proceed button cannot be clicked even though i made this choice
Left-censored data estimation
LoD (1/5 of the min. positive value)
Does anyone know how to fix this ?
(R Command History:
Save
- mSet<-InitDataObjects(“pktable”, “stat”, FALSE, 150)
- mSet<-Read.TextData(mSet, “Replacing_with_your_file_path”, “colu”, “disc”);
- mSet<-SanityCheckData(mSet)
- mSet<-PerformSanityClosure (mSet);
- mSet<-CheckContainsBlank(mSet)
- mSet<-FilterVariable(mSet, “F”, 20, “none”, -1, “mean”, 0, F,10.0)
- mSet<-PlotMissingDistr(mSet, “qc_miss_filt_0_”, “png”, 150, width=NA)
- mSet<-PlotMissingHeatmap(mSet, “qc_missheatmap_filt_0_”, “png”, 150)
- mSet<-ImputeMissingVar(mSet, method=“lod”, grpLod=F, grpMeasure=F)
)