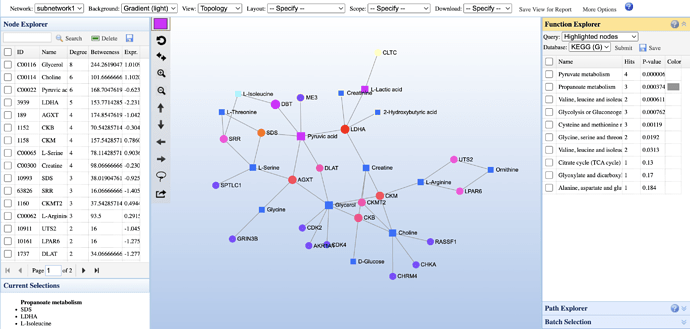
Yes. For query metabolites, you can test enriched KEGG pathways. For query genes only, you can test enriched Gene Ontologies or pathways (KEGG/Reactome).
There are three steps (please refer to the Fig. below)
-
Click the “Reset” icon
 in the network tool bar to make sure no nodes are highlighted in the current network
in the network tool bar to make sure no nodes are highlighted in the current network -
Highlight nodes based on your interest using one of the three methods:
a) Use the checkboxes in the Node Table on the left; Or
b) Use the free select icon to drag-select nodes directly on the network; Or
to drag-select nodes directly on the network; Or
c) Entering a list of node IDs to the text box in the “Batch Selection” panel on the right side -
After that, select a functional catergory from the Function Explorer section, make sure the Query option is set to Highlighted Nodes and click the Submit button.