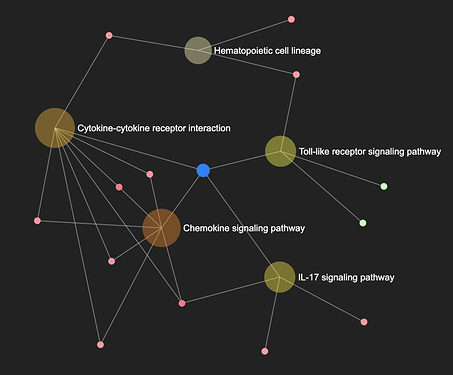
In enrichment network, gene set nodes are considered “meta-nodes” because each gene set contains gene members (child nodes). In ExpressAnalyst, double-clicking them will reveals smaller nodes that correspond to the individual genes belonging to that gene set from the analyzed gene list.
There will be an edge between the individual gene and any enriched gene set that they are a part of, so you can easily see the which genes are shared between sets. The hierarchical organization of meta-nodes allows users to customize the level of detail represented by an enrichment network.
The Fig. below shows an example generated by ExpressAnalyst, where big and semi-transparent nodes are meta-nodes (gene sets), while small and solid nodes are genes