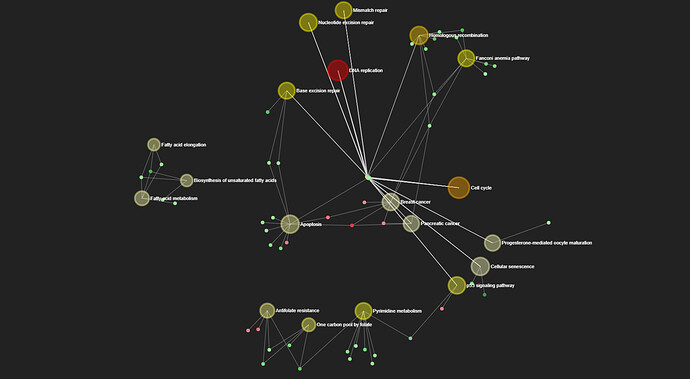
A bipartite network contains two types of nodes. In the case of enrichment network, it contains
- gene-set nodes
- gene nodes
Bipartite networks are useful to show the shared genes between gene sets. In general, if two gene sets share a lot of genes, they will be close in the enrichment network.
Please note that from the default enrichment network, you can double-click a gene-set node (which is a meta-node) to show all its genes. Refer to this post for more details on meta-node.
Below is an example of bipartite network. Larger transparent nodes represent gene sets, while smaller solid nodes represent genes (green color indicating down-regulated and red for up-regulated based on input FC values).