I’m running a series of tests using the Linear models with covariate adjustments analysis and I think there’s an error with the blocking factor function. Currently I get the same results if I have a blocking factor in the model as when I don’t have the blocking factor. I am rerunning the same models as a month ago and know the blocking factor changes the results. Thanks.
Can you provide steps and data as required for troubleshooting, or show the issue using our example data?
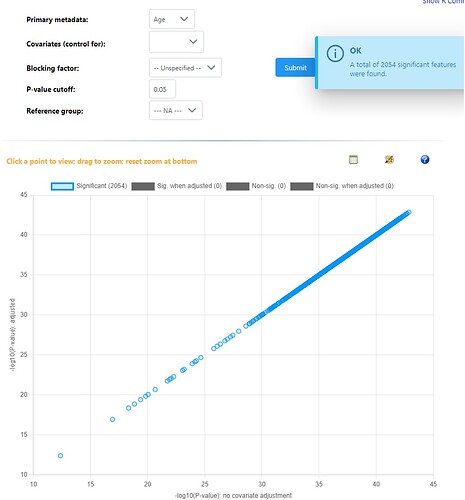
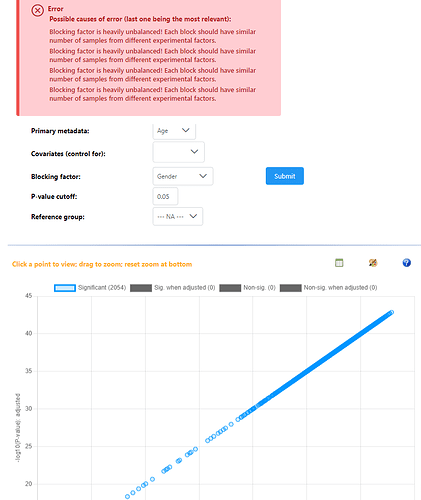
Sure. I run a simple regression model with a continuous variable as the variable of interest. I get a standard result saying all features were significant. I then add ‘site’ as a blocking factor and re-run the model and get the exact same result. This is on the same dataset I tried a month ago and not all ~450 metabolites are significant, and the blocking factor has a strong influence on the results (based on last months models).
So I think the code isn’t working at the moment…
I just tried this with the first example dataset provided and same result: every feature is significant and blocking factors/covariates don’t change the result…
Using our example dataset #1, I see clear changes with Gender as block factor. There is no related updates on the website in the past month. Have you tried to follow our Nature Protocol on this topic?
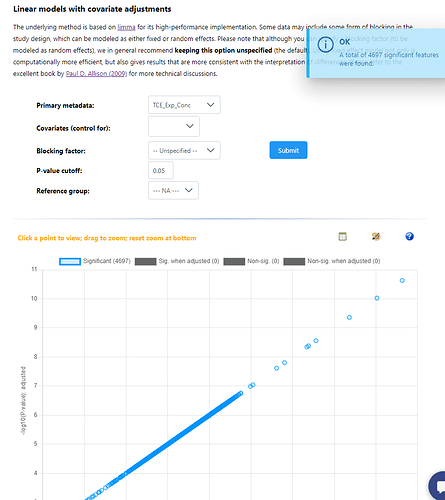
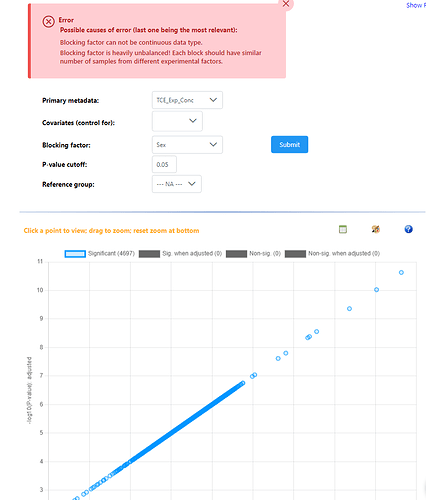
Hey, I have just used example data #1 and #2 for these screenshots. All models are saying every features is significant when compared against a single continuous variable, and all relationships are positive - which should not be possible. Then adding a blocking factor gives the exact same result, but the repeat error that blocking factors are not balanced. Again, these are very different results to what I was getting in December.
Could something be happening on my end?
Hi Damian,
I am looking into this now. I’m getting the same errors as you when the Primary Metadata is a continuous variable - it seems like its working fine for categorical primary metadata.
I’ll update you once I’ve addressed this!
Ahhh no worries, yes all my current projects are using continuous primary metadata hence my concern! Thank you!
Hello, I was just wondering if you an update on the progress of this? Thank you very much.
I just finished this up - there were a number of things to address. The changes should be online soon (probably tomorrow).
Adding the blocking factor gave an error, and the results that you saw were just the same results from the previous model (single continuous, no blocking). We were enforcing some balanced design requirements that were not compatible with continuous primary metadata - this is removed now, so using blocking factor with continuous primary metadata is now possible.
In addition, in the special case where all variables (primary and covariates) are continuous, there was a bug that has now been fixed. The number of significant results are now reasonable, and I did some stringent benchmarking comparing to other R packages to make sure it’s all working correctly.
Are you sure that it was the exact same model as several months ago? There has been no change to the code that I just modified for quite some time, so you should have gotten the same results with all continuous variables.
Thanks Jess,
The primary models I want to run at continuous metadata and a single blocking factor - so hopefully this fix will see a different result and not every feature showing a significant pattern. I will try next week to see how it runs.
The models I am running again are identical, same individuals, same metadata, same blocking factor etc. Only I have applied more QC to my dataset so there are less features and some value changes - which is why I am running them again. I’m aware this should change the results, and I remember the results for the single continuous primary metadata (no blocking) model showed a straight linear relationship with all features on the display plot, but only a top percentage were blue and ‘significant’. If this makes sense?
Thanks for addressing and I’ll update the outcomes next week!
Just letting you know the models are running fine now (both example data and my data). Thanks for fixing!
Great, glad that it’s working for you!
You might notice that we also added a new interface for performing specific contrasts for categorical primary metadata. It didn’t impact any of the testing we did with the example datasets.
Hello,
I’m attempting to use these linear models again and receiving the error message
“Error
Possible causes of error (last one being the most relevant):
Unknown Error Occurred”
when I try to use a continuous variable as the primary metadata. I’m receiving this same error when I put any variable in the covariate or blocking factor as well…