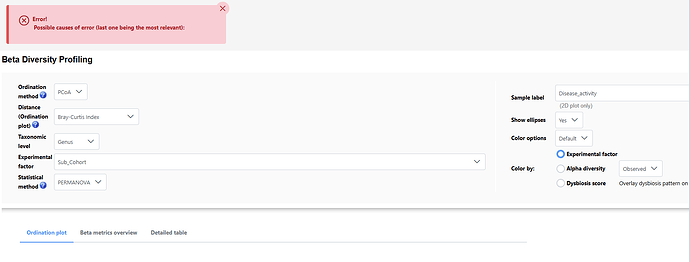
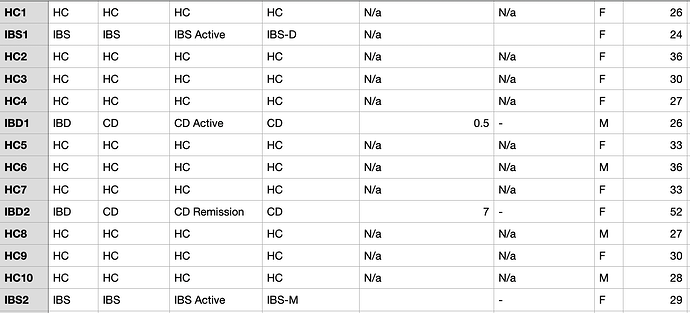
When analysing beta diversity, it comes up with this error when I try to colour by ‘experimental factor’. Do you know why please?
Thanks Jeff, apologies
- Statistical meta-analysis > Diversity meta-analysis: Beta diversity
- data attached
- PCoA, Bray Curtis, Genus, Sub-cohort, PERMANOVA
MAmetadata_to use_with activity.txt (60.5 KB)
MAtaxonomy_to use.txt (28.6 KB)
MicrobiomeAnalyst_OTU_FA_to use.txt (45.6 KB)
Also, comes up with this message when I attempt Statistical meta-analysis > Diversity meta-analysis: Alpha diversity with same data?
I am not sure why you choose to use “statistical meta-analysis” which is designed to compare data from multiple independent studies. You can see the example dataset for its accepted data format. Try to read and follow our Nature Protocol.
In addition, clean the metadata table - empty, -, N/a - all these will certainly lead to unexpected error - as computer will have no idea
Hi Jeff, thanks for your response. Oh, I see - I will have a read through of that paper, thank you.
So which setting should I be using then to process my data? I am looking for outputs such as observed OTUs, Shannon diversity, jaccard/bray-curtis plots, abundances, etc to compare mycobiome between cohorts…?
Thanks in advance
These questions are all covered in our publications. You can also attend our Omics Data Science course to get detailed training.
OK, thank you - I believe I need MDP.
Am I incorrect to have genera e.g. ‘Candida’ in the first column of my OTU/ASV and taxonomy tables?
Should it be the taxa levels separated by semicolons e.g. ‘Eukaryota; Ascomycota; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Candida’ instead or does it not matter please?
Thanks
Grace
This topic was automatically closed after 41 hours. New replies are no longer allowed.