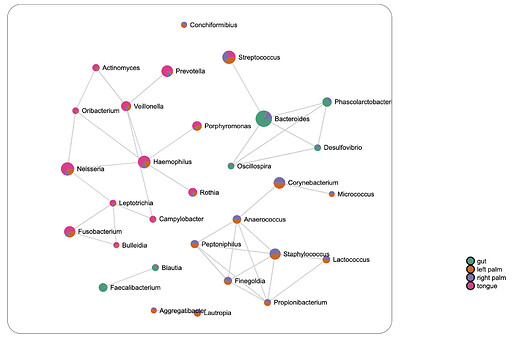
There exist several simple methods for computing correlation networks such as Pearson’s correlation, which determines if linear relationships exist between two taxa, or Spearman’s and Kendall’s rank correlation, which measures rank relationships between pairs. However, these naïve-methods often fail to address the compositional nature of microbiome data and can be unreliable due to the identification of spurious correlations. An example output from Pearson’s correlation is shown below.
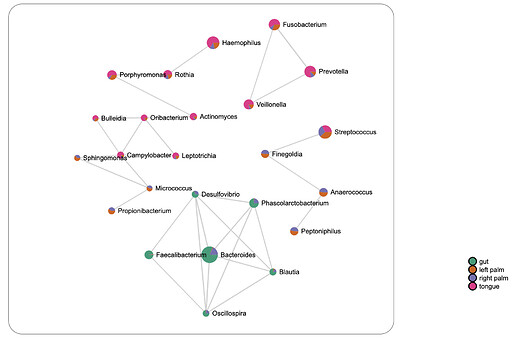
Alternatively, compositionally-robust methods including SparCC and SPIEC-EASI have been introduced, both of which make a strong assumption of a sparse correlation network. SparCC uses a log-ratio transformation and performs iterations to identify taxa pairs that are outliers to background correlations. Meanwhile, SPIEC-EASI uses graphical network models to infer the entire correlation network at once. Both methods are computationally intensive, though an efficient implementation of the SparCC algorithm named FastSpar was recently published. Due to its significantly enhanced performance, MicrobiomeAnalyst implements FastSpar as well as Pearson’s, Spearman’s and Kendall’s correlation for network creation. An example output based on SparCC using the same parameter is shown below (note it is “sparser” compared to the network above by Pearson’s method)
For more details, please refer to these papers by Friedman and Alm 2012 and Watts et al. 2019.