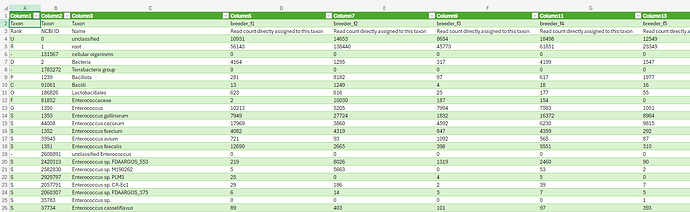
Hi everyone,
I am using MicrobiomeAnalyst to analyze OTU tables that I obtained from BugSeq. The BugSeq output for 16S data included taxonomy in the appropriate format, but I also have metagenomic whole genome sequencing data outputs that unfortunately are formatted differently. I have attached a screen shot of a portion of the OTU table I have. I am trying to figure out if there is a way to properly format it for input into MicrobiomeAnalyst, without having to manually edit the taxonomic labels (adding the full taxonomic classification with colons in between).
I’m wondering if anyone has used this type of output from BugSeq in MicrobiomeAnalyst before that can help me out!
I did some search - it seems that BugSeq is a commercial service. The format is not in common community standard format.
It makes more sense to ask the service provider to follow the community standard (which is open and stable), rather than asking a community tool to support a commercial, maybe proprietary format.
This topic was automatically closed after 9 days. New replies are no longer allowed.