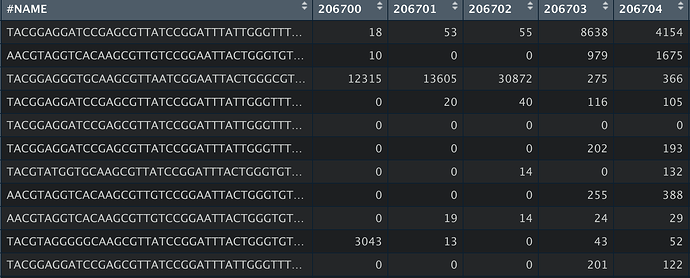
Hello! I am trying to run Tax4Fun in MicrobiomeAnalyst, but am running into trouble with my table formatting. Through the qiime2 pathway, I have an OTU, metadata and taxonomic table formatted like so:
ft_google.csv (141.3 KB)
metadata_google.csv (603 Bytes)
taxonomy_google.csv (271.4 KB)
Based on a response to a previous post, I understand that I will need to reformat my OTU table so it looks like this example:
However, I cannot figure out how to merge my sequence files from the qiime2 pathway with the OTU table so it looks like that. Any suggestions would be much appreciated!