Hi,
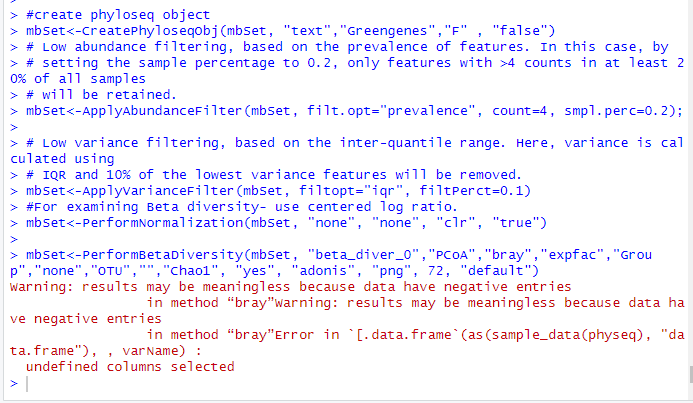
I’m trying to reproduce what the MicrobiomeAnalyst webtool produces locally using the R package- however, I cannot get the beta-diversity code to work. I’m using the IBD example dataset, and am getting these errors (using this code below, copied from the web tool).
All my packages are installed correctly, but I am getting a warning:

My filtering and normalization steps produce no errors, but then I get this error for beta diversity:
I’m not sure what is wrong with the code, especially since it seems to work within the web tool? Thank you!
Best,
Claire
| 1. |
mbSet<-Init.mbSetObj() |
| 2. |
mbSet<-SetModuleType(mbSet, “mdp”) |
| 3. |
mbSet<-Read16SAbundData(mbSet, “ibd_asv_table.txt”,“text”,“Greengenes”,“F”,“false”); |
| 4. |
mbSet<-ReadSampleTable(mbSet, “ibd_meta.csv”); |
| 5. |
mbSet<-Read16STaxaTable(mbSet, “ibd_taxa.txt”); |
| 6. |
mbSet<-ReadTreeFile(mbSet, “ibd_tree.tre”, “ibd_asv_table.txt”,“mdp”); |
| 7. |
mbSet<-SanityCheckData(mbSet, “text”); |
| 8. |
mbSet<-SanityCheckSampleData(mbSet); |
| 9. |
mbSet<-SetMetaAttributes(mbSet, “1”) |
| 10. |
mbSet<-PlotLibSizeView(mbSet, “norm_libsizes_0”,“png”); |
| 11. |
mbSet<-CreatePhyloseqObj(mbSet, “text”,“Greengenes”,“F” , “false”) |
| 12. |
mbSet<-ApplyAbundanceFilter(mbSet, “prevalence”, 4, 0.2); |
| 13. |
mbSet<-ApplyVarianceFilter(mbSet, “iqr”, 0.1); |
| 14. |
mbSet<-PerformNormalization(mbSet, “none”, “none”, “clr”, “true”); |
| 15. |
mbSet<-PerformBetaDiversity(mbSet, “beta_diver_0”,“PCoA”,“bray”,“expfac”,“Group”,“none”,“OTU”,“”,“Chao1”, “yes”, “adonis”, “png”, 72, “default”); |
| 16. |
mbSet<-PCoA3D.Anal(mbSet, “PCoA”,“bray”,“OTU”,“expfac”,“Group”,“”,“Chao1”,“beta_diver3d_0.json”) |
| 17. |
mbSet<-PerformCategoryComp(mbSet, “OTU”, “adonis”,“bray”,“Group”); |
Session info:
sessionInfo()
R version 4.3.0 (2023-04-21 ucrt)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows 10 x64 (build 19045)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.utf8 LC_CTYPE=English_United States.utf8
[3] LC_MONETARY=English_United States.utf8 LC_NUMERIC=C
[5] LC_TIME=English_United States.utf8
time zone: America/Los_Angeles
tzcode source: internal
attached base packages:
[1] grid stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] MicrobiomeAnalystR_2.0 devtools_2.4.5
[3] usethis_2.1.6 lubridate_1.9.2
[5] forcats_1.0.0 purrr_1.0.1
[7] readr_2.1.4 tidyr_1.3.0
[9] tibble_3.2.1 tidyverse_2.0.0
[11] memoise_2.0.1 dplyr_1.1.2
[13] qs_0.25.5 ppcor_1.1
[15] ggrepel_0.9.3 viridis_0.6.3
[17] viridisLite_0.4.2 R.utils_2.12.2
[19] R.oo_1.25.0 R.methodsS3_1.8.2
[21] globaltest_5.54.0 survival_3.5-5
[23] edgeR_3.42.2 splitstackshape_1.4.8
[25] gridExtra_2.3 ape_5.7-1
[27] stringr_1.5.0 reshape_0.8.9
[29] data.table_1.14.8 genefilter_1.82.1
[31] xtable_1.8-4 pheatmap_1.0.12
[33] ggfortify_0.4.16 RJSONIO_1.3-1.8
[35] vegan_2.6-4 lattice_0.21-8
[37] permute_0.9-7 DESeq2_1.40.1
[39] SummarizedExperiment_1.30.1 MatrixGenerics_1.12.0
[41] matrixStats_0.63.0 GenomicRanges_1.52.0
[43] GenomeInfoDb_1.36.0 IRanges_2.34.0
[45] S4Vectors_0.38.1 MASS_7.3-58.4
[47] metagenomeSeq_1.42.0 glmnet_4.1-7
[49] Matrix_1.5-4 limma_3.56.1
[51] Biobase_2.60.0 BiocGenerics_0.46.0
[53] randomForest_4.7-1.1 BiocParallel_1.34.2
[55] igraph_1.4.3 Cairo_1.6-0
[57] gplots_3.1.3 ggplot2_3.4.2
[59] RColorBrewer_1.1-3 biomformat_1.28.0
[61] pryr_0.1.6 metacoder_0.3.6
[63] phyloseq_1.44.0 pacman_0.5.1
loaded via a namespace (and not attached):
[1] splines_4.3.0 later_1.3.1 bitops_1.0-7
[4] XML_3.99-0.14 lifecycle_1.0.3 rprojroot_2.0.3
[7] processx_3.8.1 magrittr_2.0.3 rmarkdown_2.21
[10] yaml_2.3.7 remotes_2.4.2 httpuv_1.6.11
[13] Wrench_1.18.0 sessioninfo_1.2.2 pkgbuild_1.4.0
[16] DBI_1.1.3 ade4_1.7-22 pkgload_1.3.2
[19] zlibbioc_1.46.0 RCurl_1.98-1.12 GenomeInfoDbData_1.2.10
[22] annotate_1.78.0 codetools_0.2-19 DelayedArray_0.26.3
[25] RApiSerialize_0.1.2 tidyselect_1.2.0 shape_1.4.6
[28] jsonlite_1.8.4 multtest_2.56.0 ellipsis_0.3.2
[31] iterators_1.0.14 foreach_1.5.2 tools_4.3.0
[34] Rcpp_1.0.10 glue_1.6.2 xfun_0.39
[37] mgcv_1.8-42 withr_2.5.0 BiocManager_1.30.20
[40] fastmap_1.1.1 rhdf5filters_1.12.1 fansi_1.0.4
[43] callr_3.7.3 caTools_1.18.2 digest_0.6.31
[46] timechange_0.2.0 R6_2.5.1 mime_0.12
[49] colorspace_2.1-0 gtools_3.9.4 RSQLite_2.3.1
[52] utf8_1.2.3 generics_0.1.3 prettyunits_1.1.1
[55] httr_1.4.6 htmlwidgets_1.6.2 S4Arrays_1.0.4
[58] Tax4Fun_0.3.1 pkgconfig_2.0.3 gtable_0.3.3
[61] blob_1.2.4 XVector_0.40.0 htmltools_0.5.5
[64] profvis_0.3.8 scales_1.2.1 png_0.1-8
[67] knitr_1.43 rstudioapi_0.14 tzdb_0.4.0
[70] reshape2_1.4.4 curl_5.0.0 nlme_3.1-162
[73] cachem_1.0.8 rhdf5_2.44.0 KernSmooth_2.23-20
[76] parallel_4.3.0 miniUI_0.1.1.1 AnnotationDbi_1.62.1
[79] desc_1.4.2 pillar_1.9.0 vctrs_0.6.2
[82] urlchecker_1.0.1 promises_1.2.0.1 stringfish_0.15.7
[85] cluster_2.1.4 evaluate_0.21 cli_3.6.1
[88] locfit_1.5-9.7 compiler_4.3.0 rlang_1.1.1
[91] crayon_1.5.2 ps_1.7.5 plyr_1.8.8
[94] fs_1.6.2 stringi_1.7.12 munsell_0.5.0
[97] Biostrings_2.68.1 hms_1.1.3 taxa_0.4.2
[100] bit64_4.0.5 Rhdf5lib_1.22.0 KEGGREST_1.40.0
[103] shiny_1.7.4 RcppParallel_5.1.7 bit_4.0.5