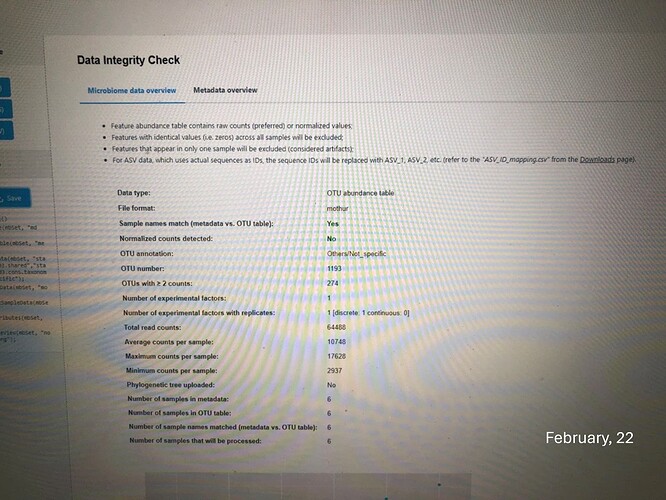
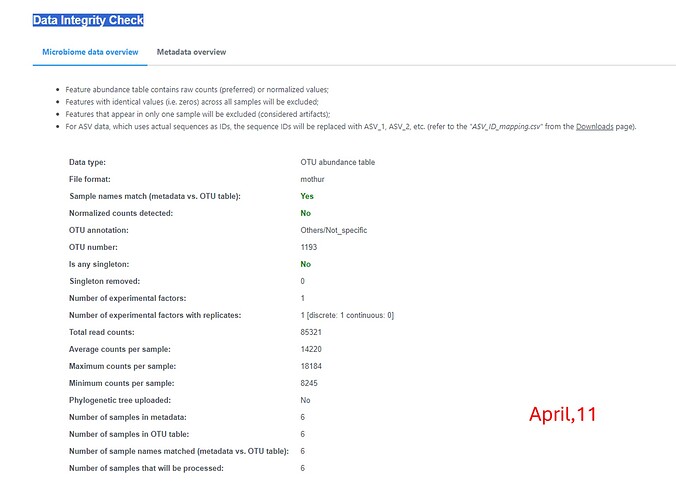
I notice that my relative abundance graphs (phylum and genus) as well as the data integrity check are different from those obtained in february (last analysis), though I used the same OTU table, metadata file and consensus taxonomy files which were generated by Mothur. Is this normal? Thanks
Hello,
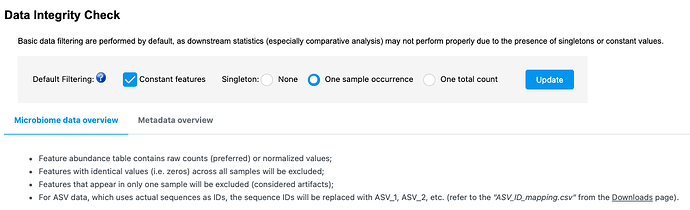
Thanks for the message. This is because we have update the default pre-processing step recently. Previously, we remove the features present in only one sample which are considered as artifact and the constant features which are meaningless for differential analysis. To enhance the performance, we now allow users to customize the default processing based on their dataset. You can find more information on the updated web page::
If you stick with the default option, you will be able to reproduce the results generated in February.
Hope this helps!
Thank you for your reply ![]()