Hi,
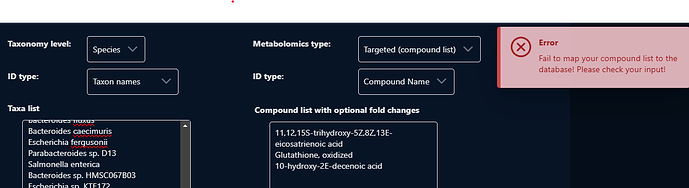
I’m using Microbial Metabolomics (co-analysis). I have put my species of bacteria and compound names (metabolites) in Feature lists tap. But I keep getting an error as attached image and I think those 3 metabolites below are causing error. I have checked with your Omics assistant but didn’t work. It would be appreciated if you can check my compound name.
11,12,15S-trihydroxy-5Z,8Z,13E-eicosatrienoic acid
Glutathione, oxidized
10-hydroxy-2E-decenoic acid
Thank you!