Hello,
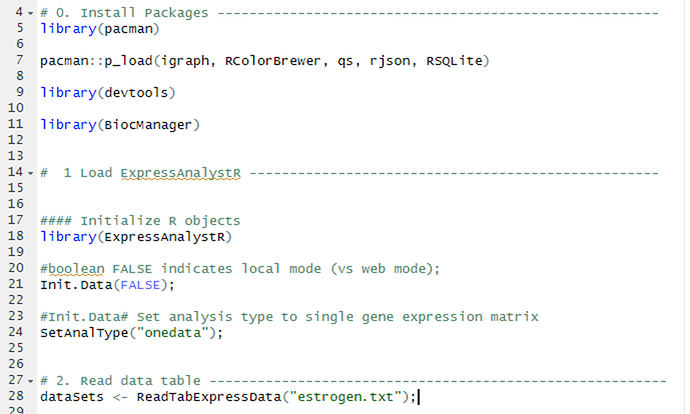
After installing the expressanalyst package in R, I wanted to run the gene expression matrix estrogen example found on the website. However, it is not running properly, and I think is related to the ReadTabExpressData function as I got this :
dataSets ← ReadTabExpressData(“estrogen.txt”);
Error in if (sum(!good.inx) > 0) { :
missing value where TRUE/FALSE needed
I attach the script and console:
After that if I run the PerformDataAnnot function I get the following:
dataSets ← PerformDataAnnot(“estrogen.txt”, “hsa”, “array”, “hgu95av2”, “mean”);
Error in qs::qread(fileName) : QS format not detected
I tried uploading the database directly, changing the format, checking the txt file, etc, but it just does not run.
I would very much appreciate any help.