Hello,
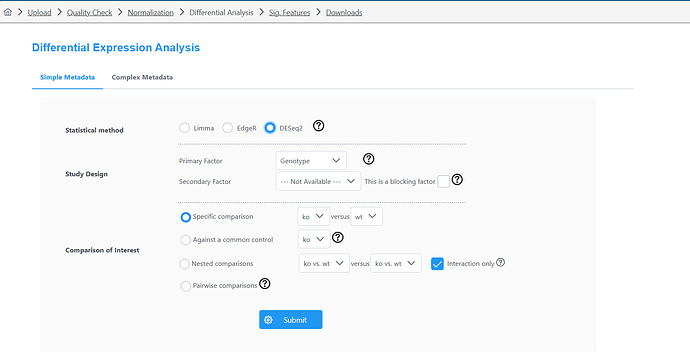
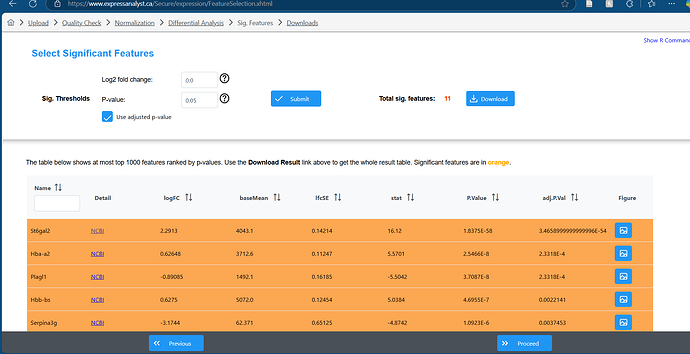
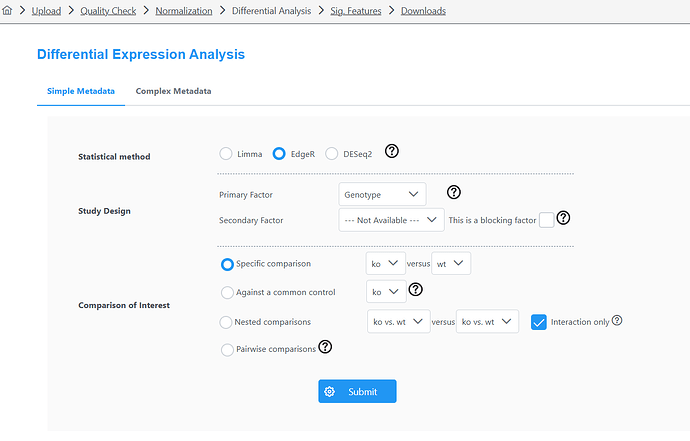
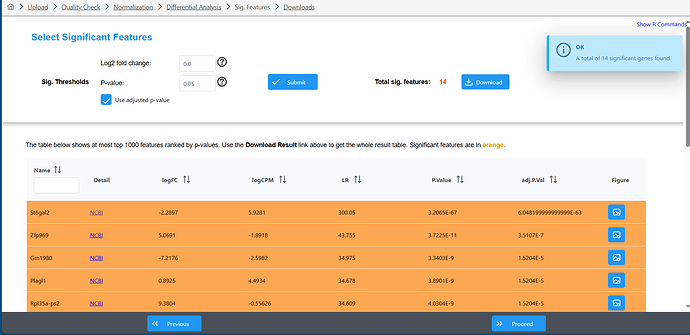
I was using express analyst today and noticed that the Edger analysis and the DESeq2 are showing opposite resultes for LogFC (I didn’t change any other parameters between the two). I have done this analysis before so I know that the DESeq2 is wrong.
I also mentioned in a previous post, and followed up through email that the ridgeline plot are not displaying gene name anymore, only EntrezID, making the navigation tool much less useful to browse through the data in a comprehensive way.
Thank you very much for the work you do on this great platform!
Jeanne