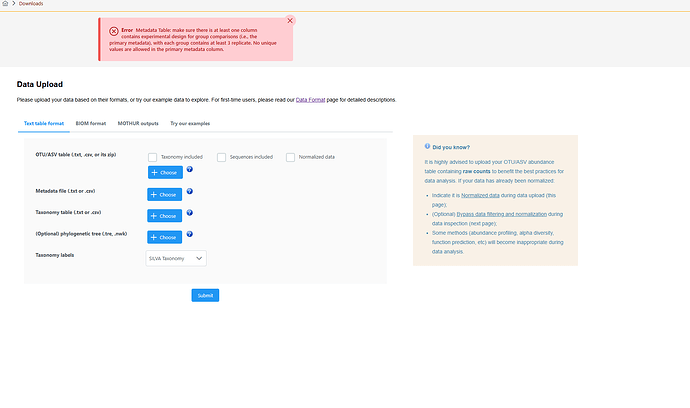
Hi Microbiome analyst Team,
I am analyz
Galaxy258-[Galaxy237-[0.03].mothur.cons.taxonomy].csv (52.6 KB)
Galaxy256-[234__Classify.otu_on_data_148,_data_150,_and_data_231__tax.summary.zip] (1).csv (6.0 KB)
ing the 16s metagenomics data (Sequenced by Illumina Miseq, paired end, V2 kit), I have used Galaxy Mothur tool for analyzing my data. I have the files in csv/txt format. I read the guidelines on the data format page and tried to followed to best. But, Its displaying the error. I need to use this tool for visualizing the data.
If your team can help me troubleshotting this issue, I have attached the files for reference.
Thanks,
Shweta
Metatable.txt (94 Bytes)